rm(list=ls())
library(TAM)
library(WrightMap)
library(tidyverse)
# load in the dataset
responses <- read_csv('data/pc-data.csv')
# drop the first six columns
resp <- responses %>%
select(-c(1:6))
# choose the columns that start with C1_
resp_c1 <- resp %>%
select(starts_with('C1_'))
mod1 <- tam.jml(resp_c1)
thres <- tam.threshold(mod1) # find Thurstonian thresholds28 Fitting the Partial Credit Model with TAM
28.1 Items in section 1, C1_
wrightMap(mod1$WLE, thres, item.side = itemClassic)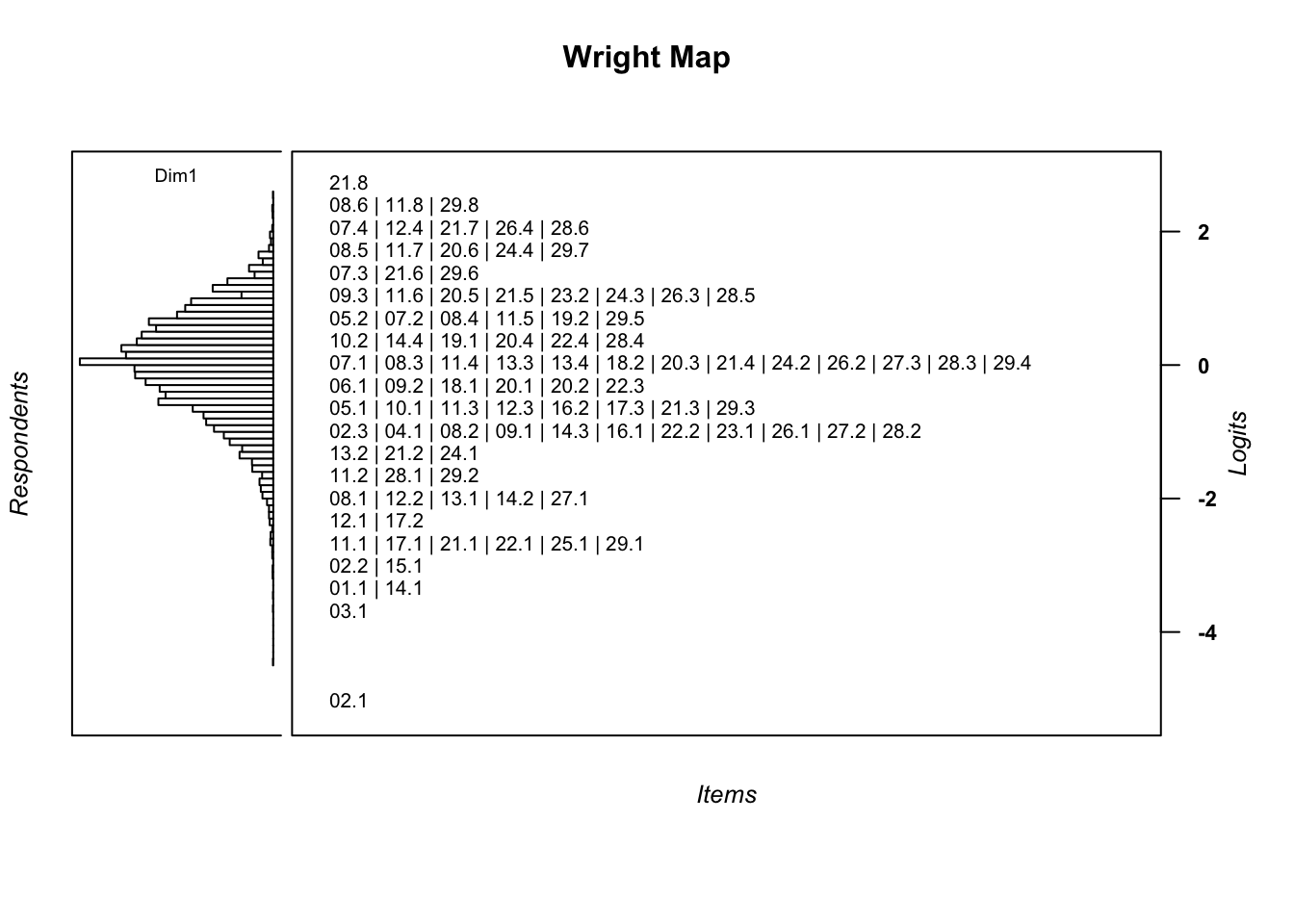
Cat1 Cat2 Cat3 Cat4 Cat5 Cat6
C1_1ai -3.49868774 NA NA NA NA NA
C1_1aii -4.97634888 -2.8871155 -1.10000610 NA NA NA
C1_1bi -3.64773560 NA NA NA NA NA
C1_1bii -0.97677612 NA NA NA NA NA
C1_1biii -0.69223022 0.7973328 NA NA NA NA
C1_1biv -0.28372192 NA NA NA NA NA
C1_1ci -0.06912231 0.5939026 1.31918335 2.14205933 NA NA
C1_1cii -2.08602905 -0.9289856 0.15719604 0.74002075 1.5264587 2.401520
C1_1di -0.96963501 -0.2681580 0.89932251 NA NA NA
C1_1dii -0.81106567 0.4144592 NA NA NA NA
C1_1e -2.71646118 -1.6004333 -0.77206421 -0.12258911 0.5169983 1.003510
C1_1eSPaG -2.39767456 -1.9955750 -0.73745728 2.14389038 NA NA
C1_2ai -2.13308716 -1.2915344 -0.02169800 -0.01730347 NA NA
C1_2aii -3.28060913 -2.0669861 -0.89144897 0.28500366 NA NA
C1_2bi -2.89389038 NA NA NA NA NA
C1_2bii -0.99819946 -0.5887756 NA NA NA NA
C1_2biii -2.85470581 -2.4703674 -0.72244263 NA NA NA
C1_2biv -0.39450073 -0.1452942 NA NA NA NA
C1_2bv 0.27548218 0.7987976 NA NA NA NA
C1_2c -0.49594116 -0.2167053 0.01748657 0.40988159 0.9381409 1.690155
C1_2d -2.56704712 -1.4623718 -0.64590454 0.02005005 0.8806458 1.355255
C1_3ai -2.69851685 -1.1644592 -0.47341919 0.27035522 NA NA
C1_3aii -0.91488647 1.0318909 NA NA NA NA
C1_3aiii -1.24703979 0.1050110 1.00900269 1.70498657 NA NA
C1_3bi -2.66152954 NA NA NA NA NA
C1_3bii -0.98007202 -0.0319519 0.91909790 1.87692261 NA NA
C1_3ci -2.12612915 -0.9866638 -0.14987183 NA NA NA
C1_3cii -1.57296753 -0.9277039 -0.11141968 0.40438843 1.0650330 1.869781
C1_3d -2.59176636 -1.5603333 -0.63272095 0.06381226 0.8009949 1.265717
Cat7 Cat8
C1_1ai NA NA
C1_1aii NA NA
C1_1bi NA NA
C1_1bii NA NA
C1_1biii NA NA
C1_1biv NA NA
C1_1ci NA NA
C1_1cii NA NA
C1_1di NA NA
C1_1dii NA NA
C1_1e 1.563629 2.297333
C1_1eSPaG NA NA
C1_2ai NA NA
C1_2aii NA NA
C1_2bi NA NA
C1_2bii NA NA
C1_2biii NA NA
C1_2biv NA NA
C1_2bv NA NA
C1_2c NA NA
C1_2d 1.944489 2.624725
C1_3ai NA NA
C1_3aii NA NA
C1_3aiii NA NA
C1_3bi NA NA
C1_3bii NA NA
C1_3ci NA NA
C1_3cii NA NA
C1_3d 1.811005 2.438873wrightMap(mod1$WLE, thres) # try different display options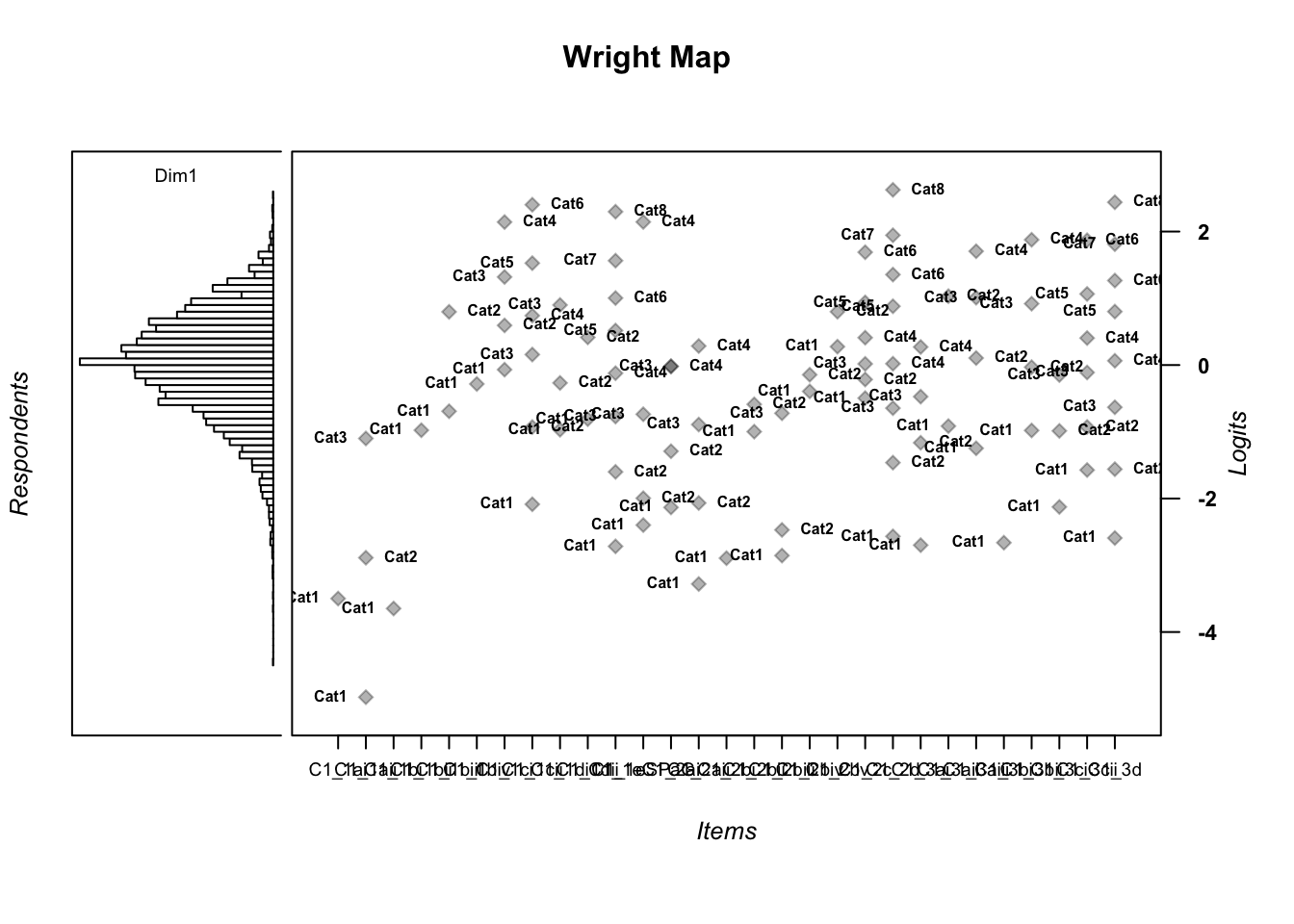
Cat1 Cat2 Cat3 Cat4 Cat5 Cat6
C1_1ai -3.49868774 NA NA NA NA NA
C1_1aii -4.97634888 -2.8871155 -1.10000610 NA NA NA
C1_1bi -3.64773560 NA NA NA NA NA
C1_1bii -0.97677612 NA NA NA NA NA
C1_1biii -0.69223022 0.7973328 NA NA NA NA
C1_1biv -0.28372192 NA NA NA NA NA
C1_1ci -0.06912231 0.5939026 1.31918335 2.14205933 NA NA
C1_1cii -2.08602905 -0.9289856 0.15719604 0.74002075 1.5264587 2.401520
C1_1di -0.96963501 -0.2681580 0.89932251 NA NA NA
C1_1dii -0.81106567 0.4144592 NA NA NA NA
C1_1e -2.71646118 -1.6004333 -0.77206421 -0.12258911 0.5169983 1.003510
C1_1eSPaG -2.39767456 -1.9955750 -0.73745728 2.14389038 NA NA
C1_2ai -2.13308716 -1.2915344 -0.02169800 -0.01730347 NA NA
C1_2aii -3.28060913 -2.0669861 -0.89144897 0.28500366 NA NA
C1_2bi -2.89389038 NA NA NA NA NA
C1_2bii -0.99819946 -0.5887756 NA NA NA NA
C1_2biii -2.85470581 -2.4703674 -0.72244263 NA NA NA
C1_2biv -0.39450073 -0.1452942 NA NA NA NA
C1_2bv 0.27548218 0.7987976 NA NA NA NA
C1_2c -0.49594116 -0.2167053 0.01748657 0.40988159 0.9381409 1.690155
C1_2d -2.56704712 -1.4623718 -0.64590454 0.02005005 0.8806458 1.355255
C1_3ai -2.69851685 -1.1644592 -0.47341919 0.27035522 NA NA
C1_3aii -0.91488647 1.0318909 NA NA NA NA
C1_3aiii -1.24703979 0.1050110 1.00900269 1.70498657 NA NA
C1_3bi -2.66152954 NA NA NA NA NA
C1_3bii -0.98007202 -0.0319519 0.91909790 1.87692261 NA NA
C1_3ci -2.12612915 -0.9866638 -0.14987183 NA NA NA
C1_3cii -1.57296753 -0.9277039 -0.11141968 0.40438843 1.0650330 1.869781
C1_3d -2.59176636 -1.5603333 -0.63272095 0.06381226 0.8009949 1.265717
Cat7 Cat8
C1_1ai NA NA
C1_1aii NA NA
C1_1bi NA NA
C1_1bii NA NA
C1_1biii NA NA
C1_1biv NA NA
C1_1ci NA NA
C1_1cii NA NA
C1_1di NA NA
C1_1dii NA NA
C1_1e 1.563629 2.297333
C1_1eSPaG NA NA
C1_2ai NA NA
C1_2aii NA NA
C1_2bi NA NA
C1_2bii NA NA
C1_2biii NA NA
C1_2biv NA NA
C1_2bv NA NA
C1_2c NA NA
C1_2d 1.944489 2.624725
C1_3ai NA NA
C1_3aii NA NA
C1_3aiii NA NA
C1_3bi NA NA
C1_3bii NA NA
C1_3ci NA NA
C1_3cii NA NA
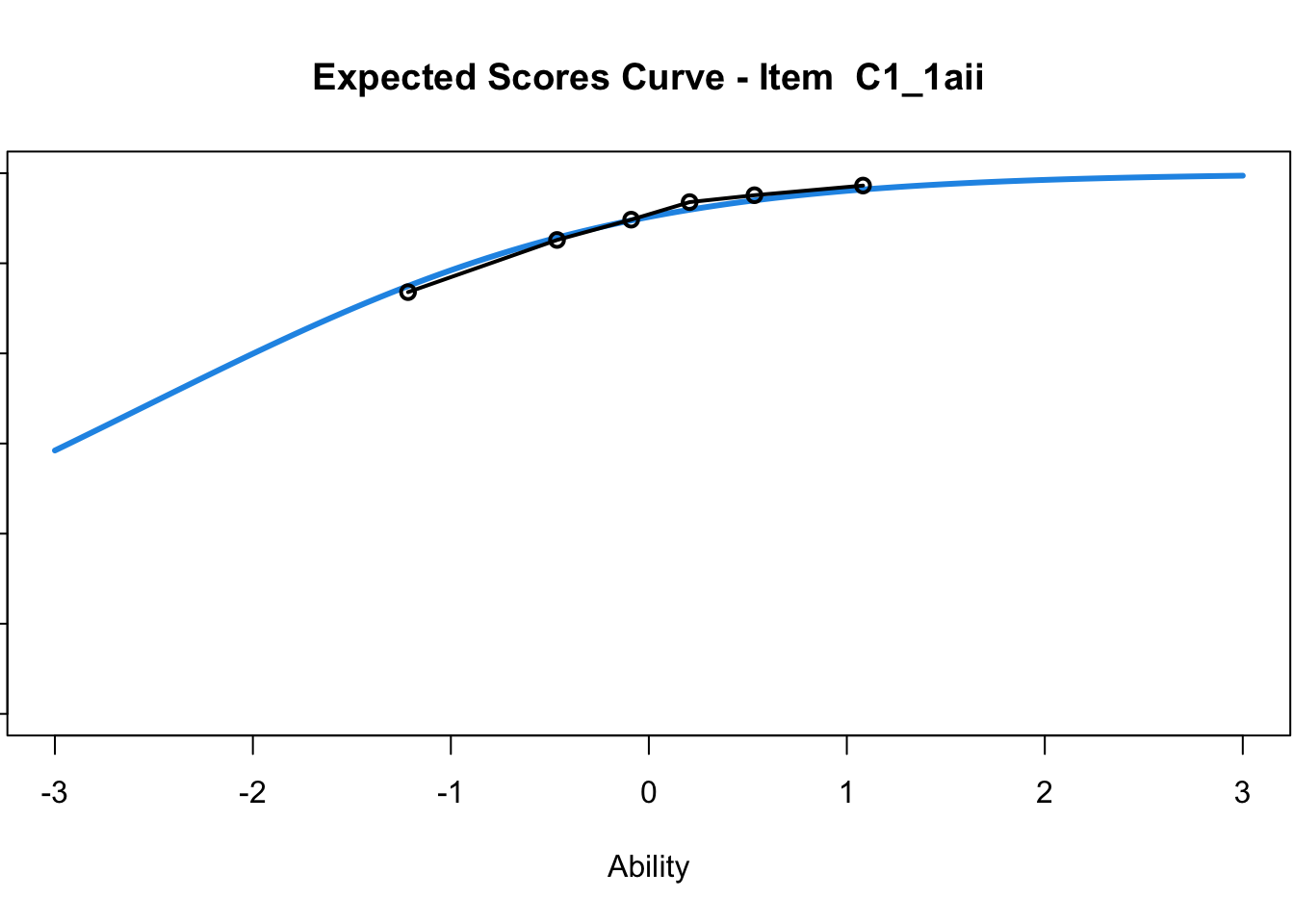
C1_3d 1.811005 2.438873plot(mod1) #Expected score curves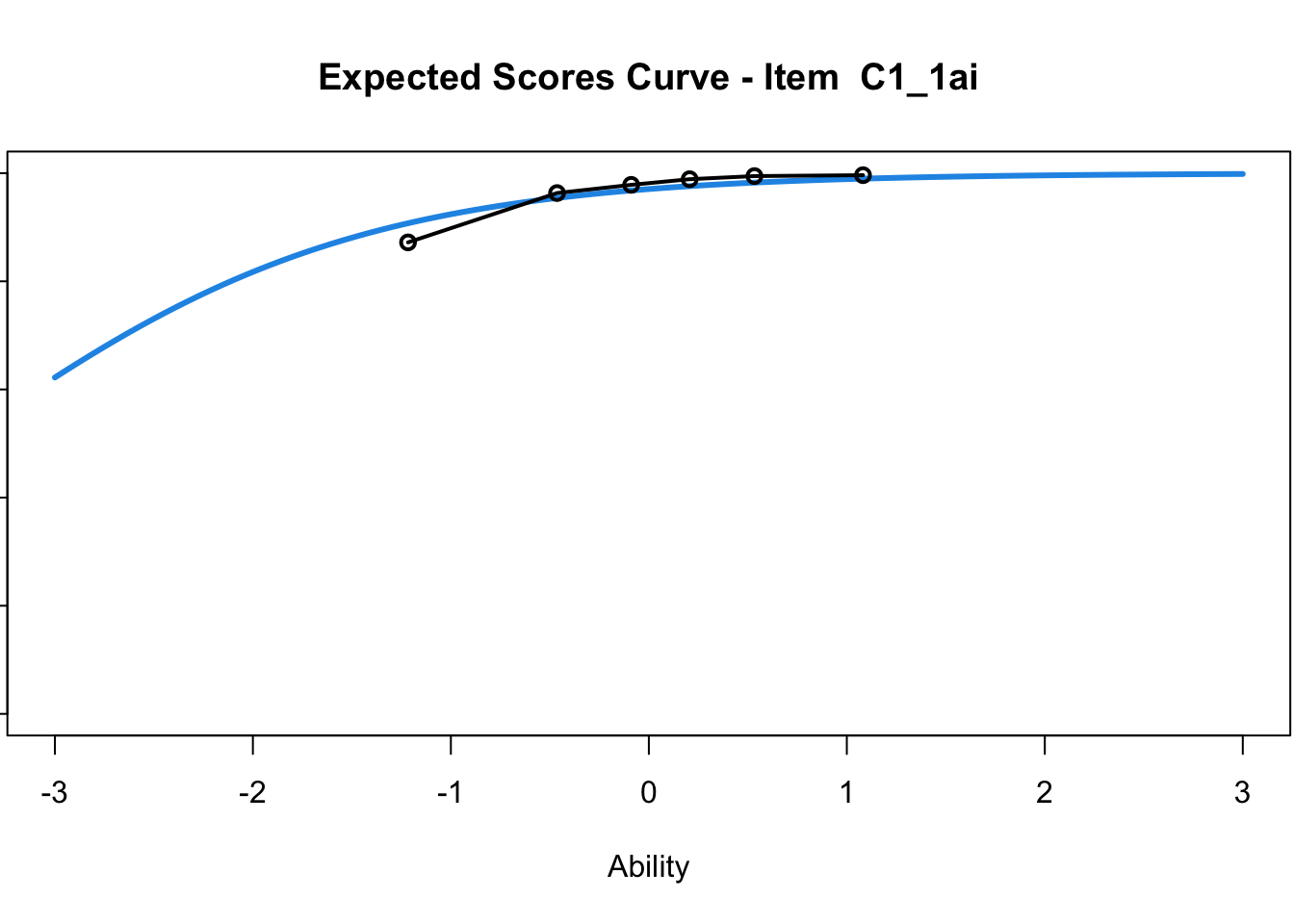
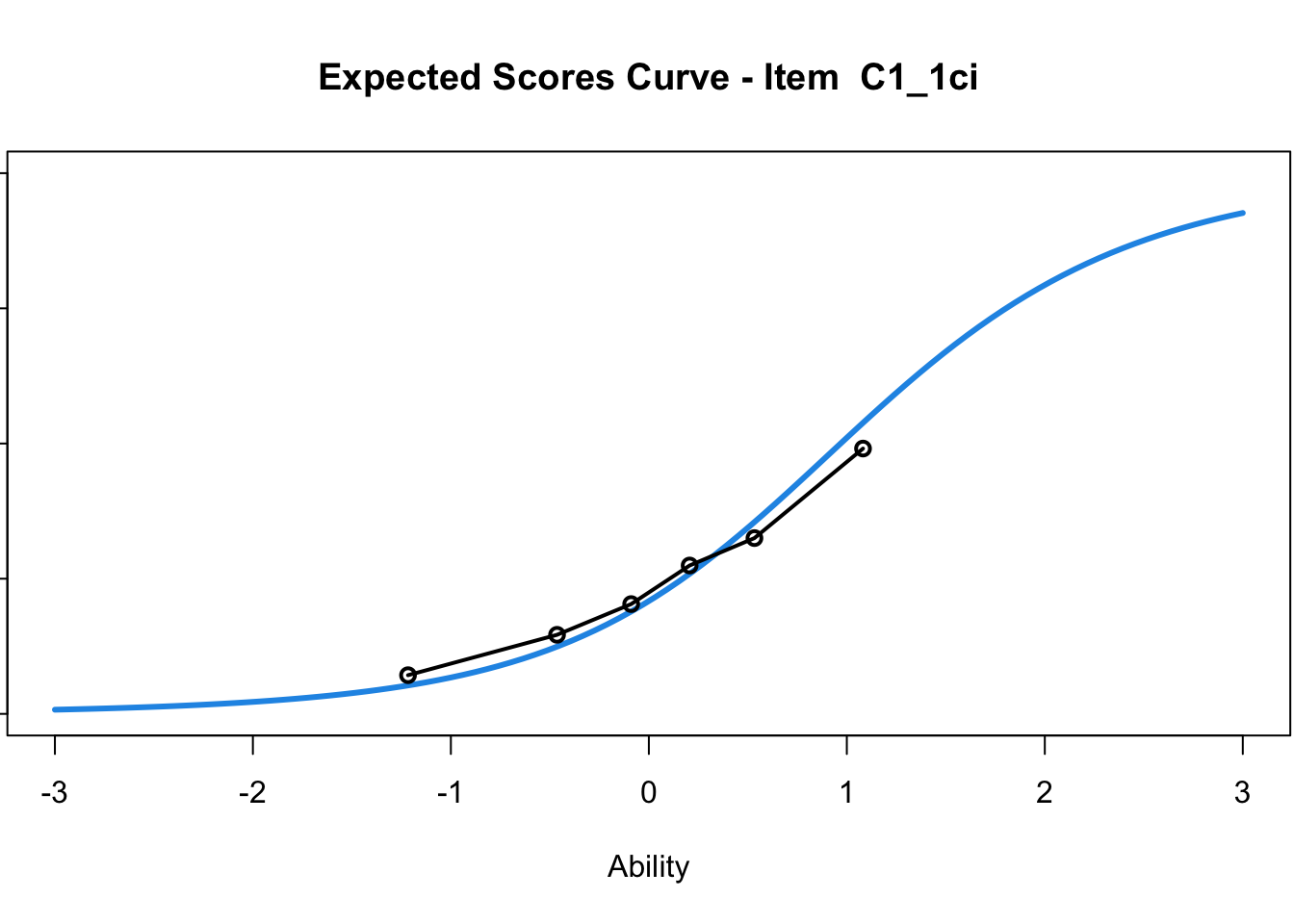
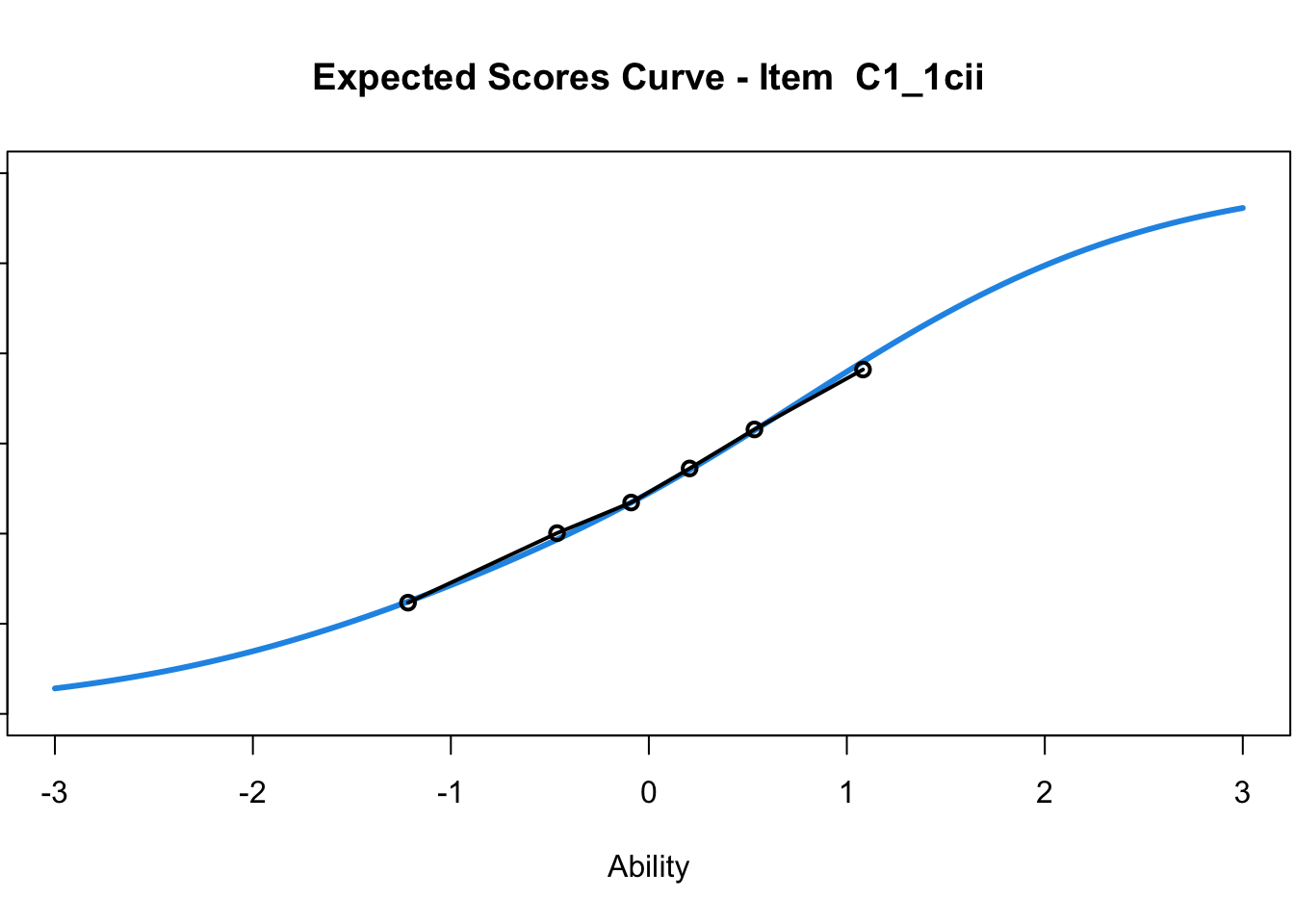
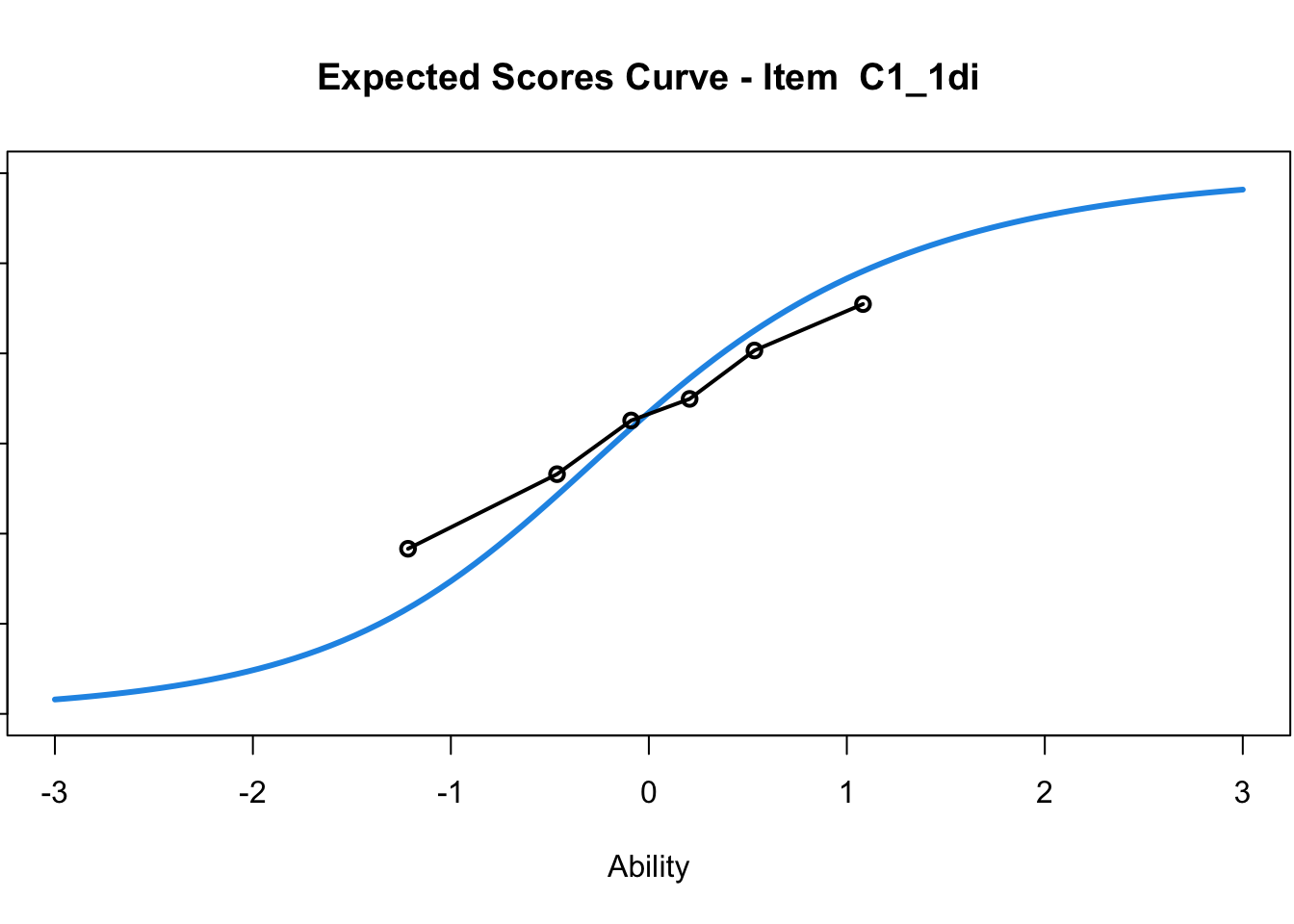
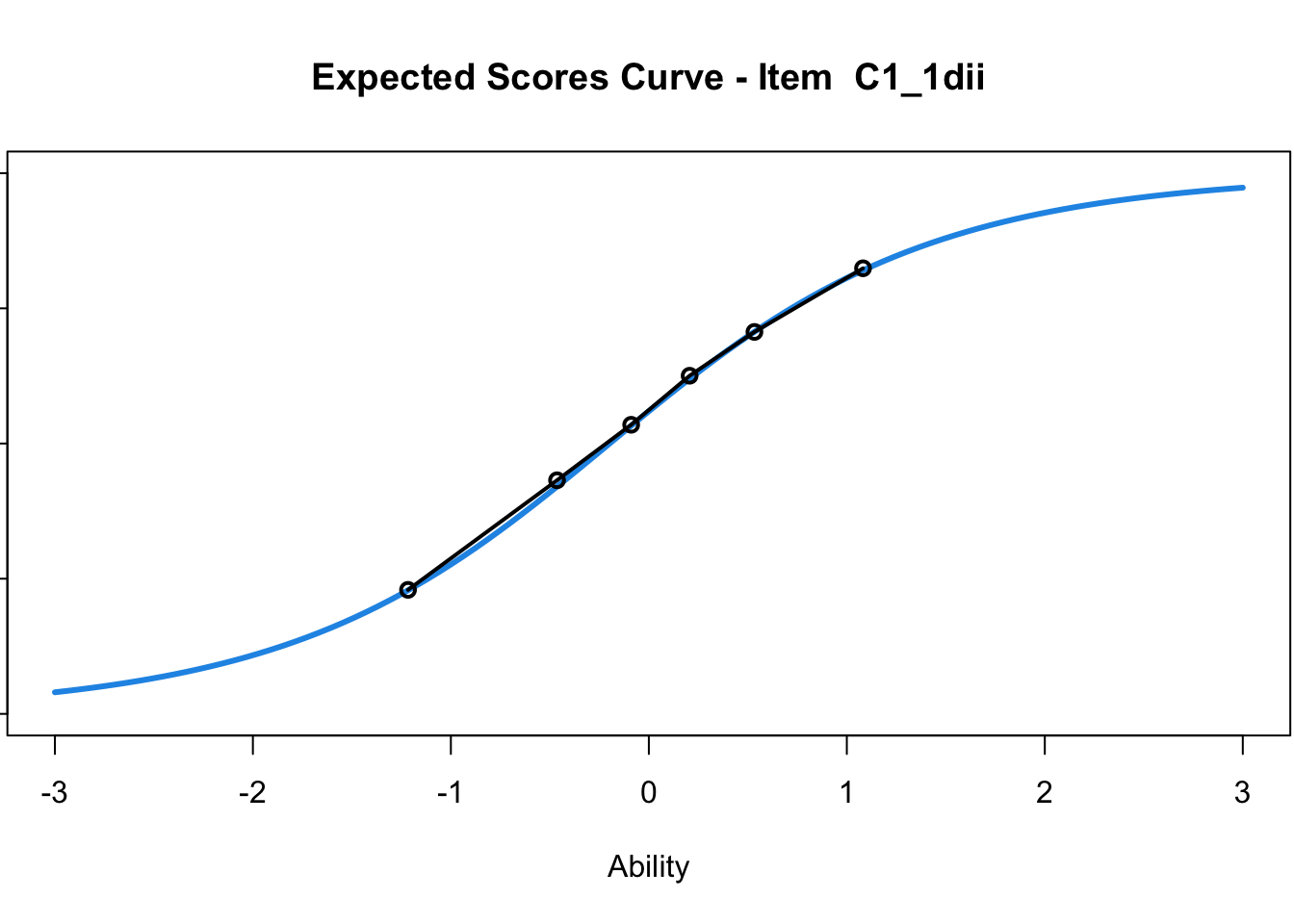
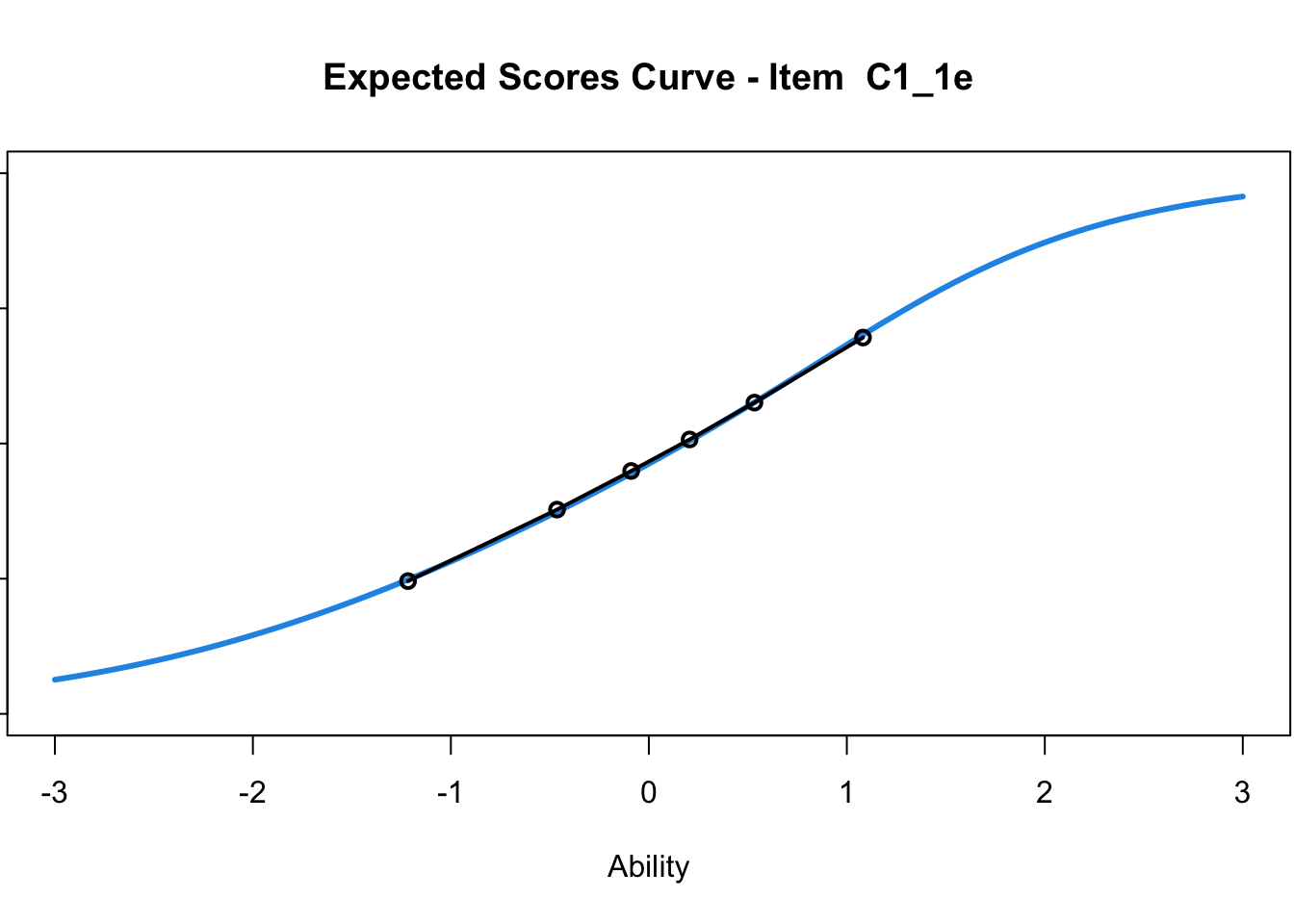
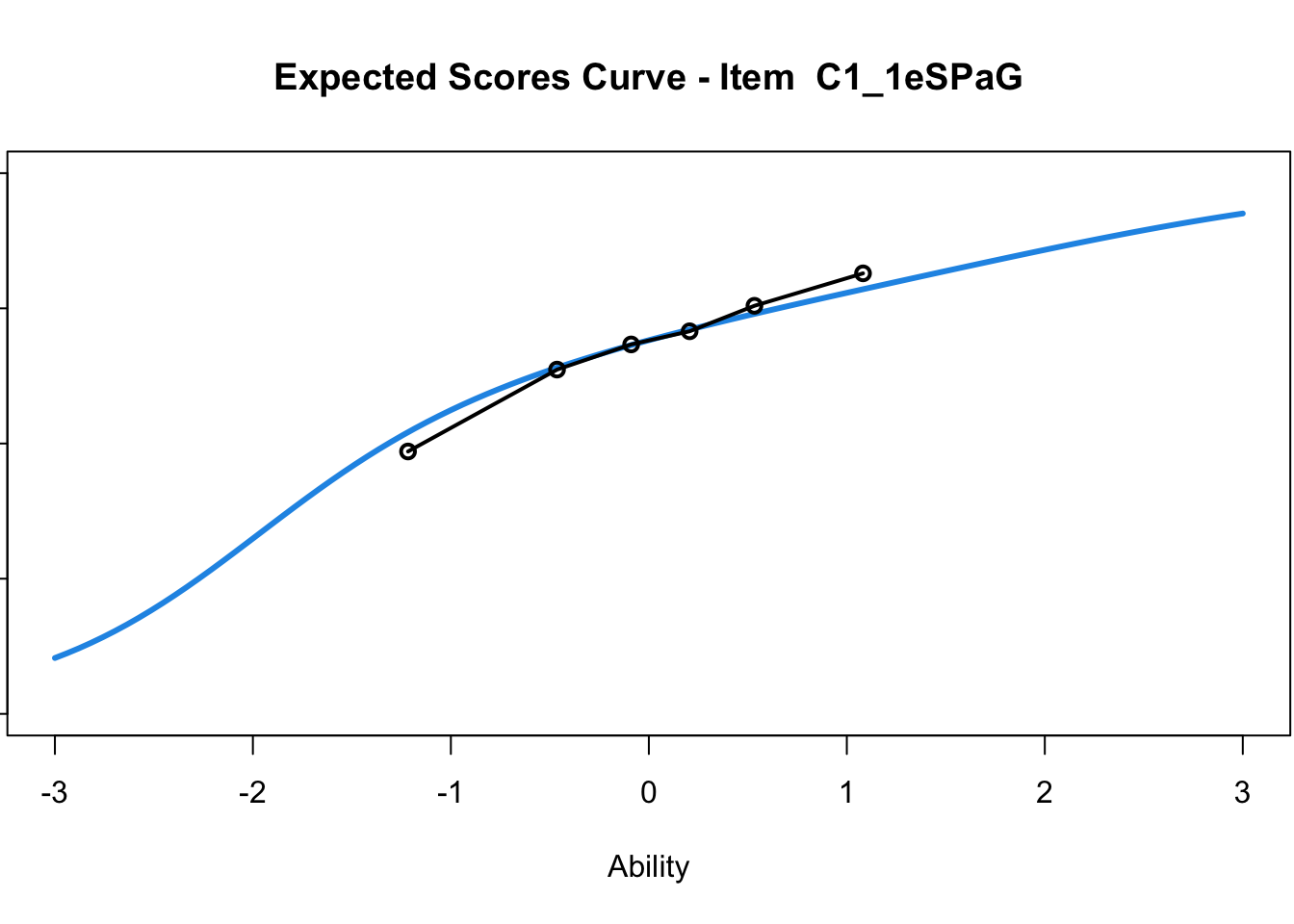
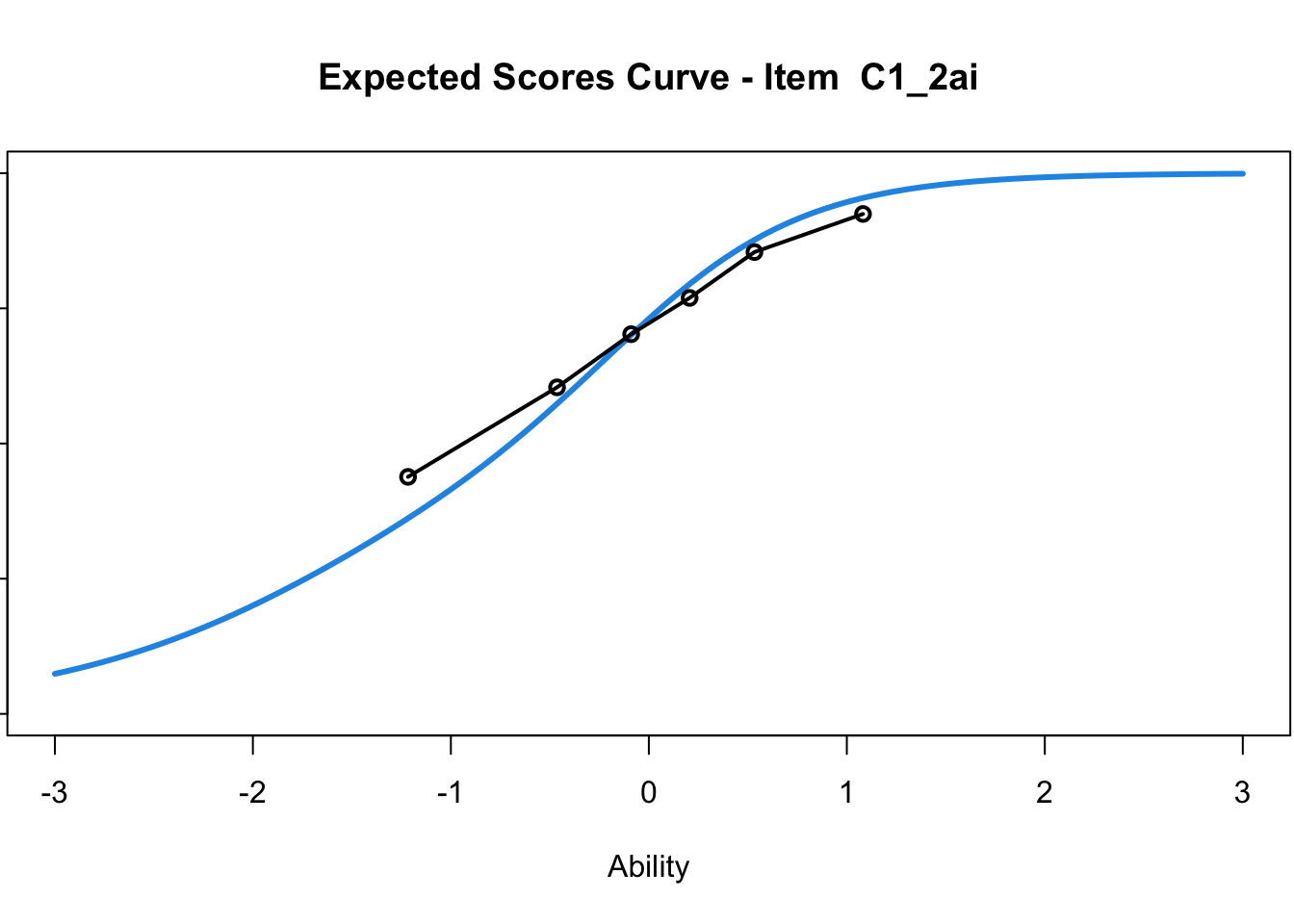
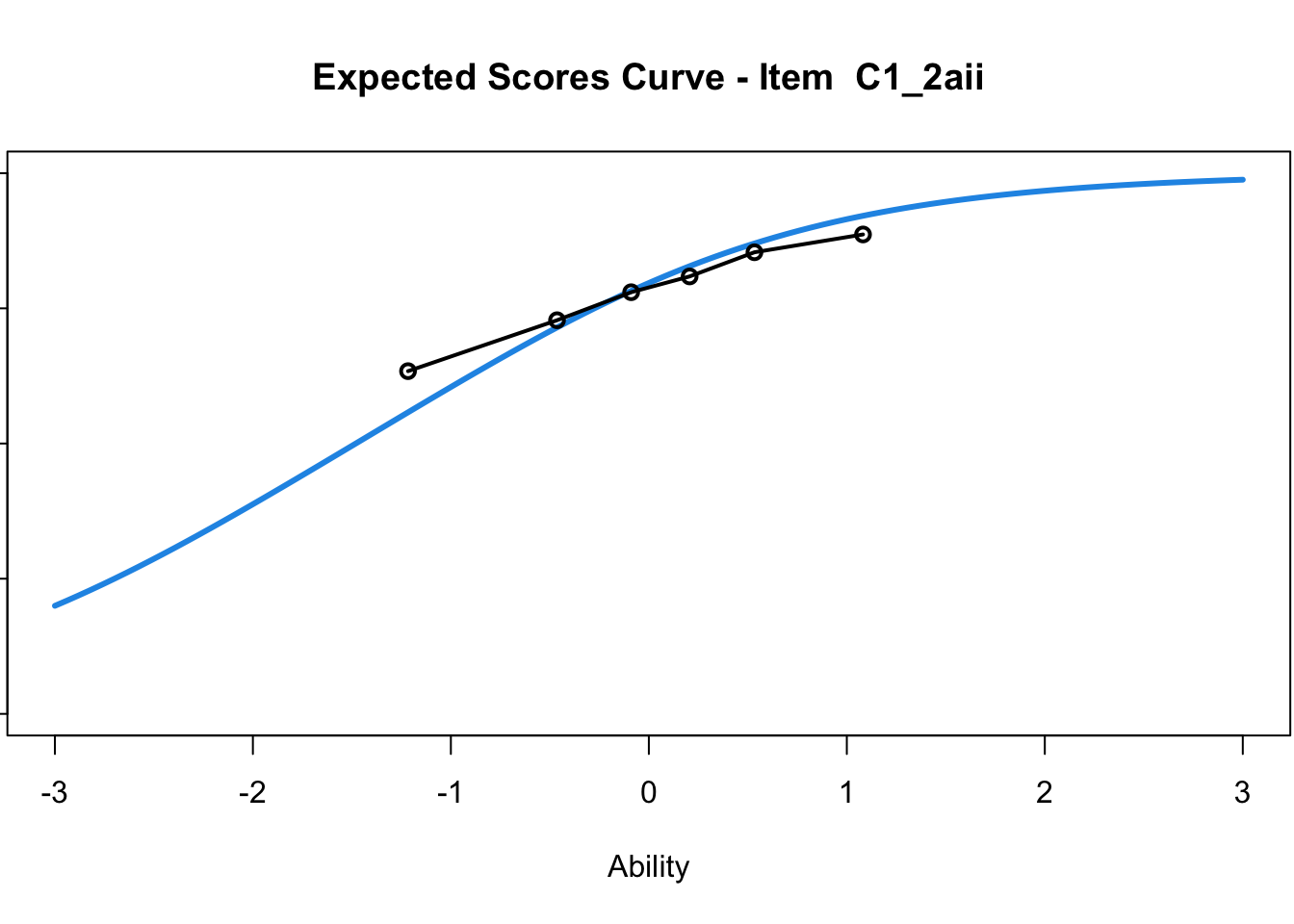
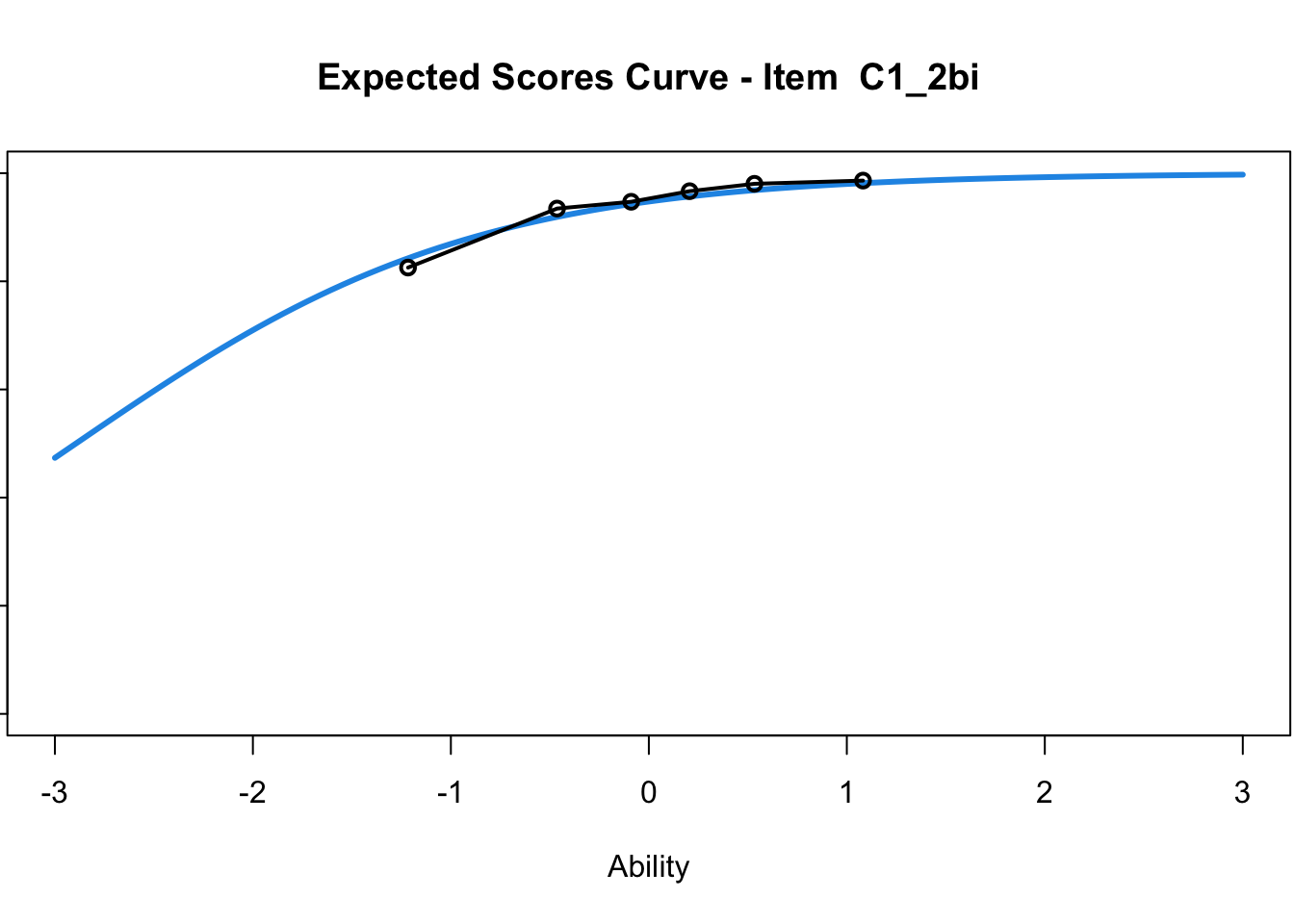
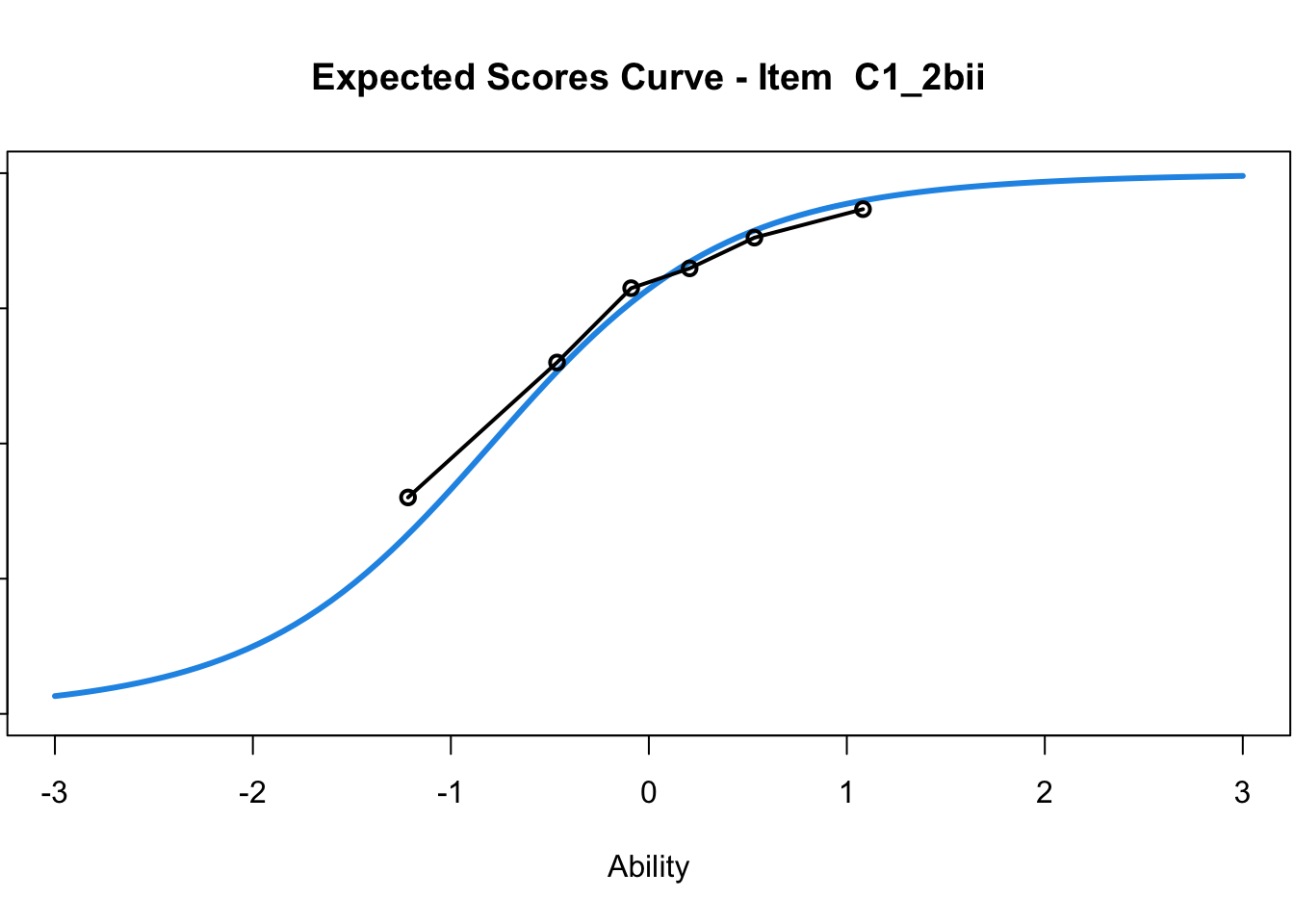
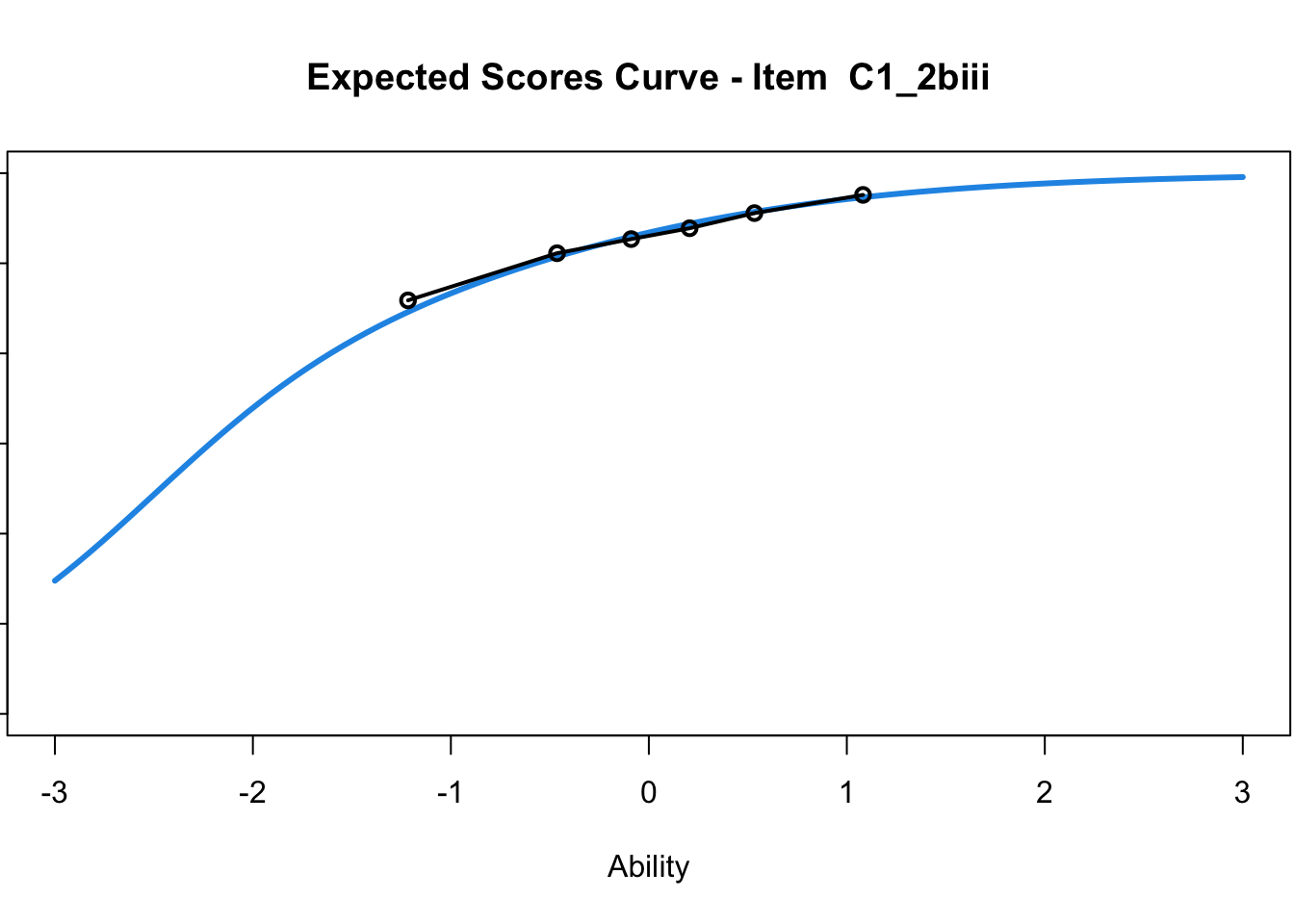
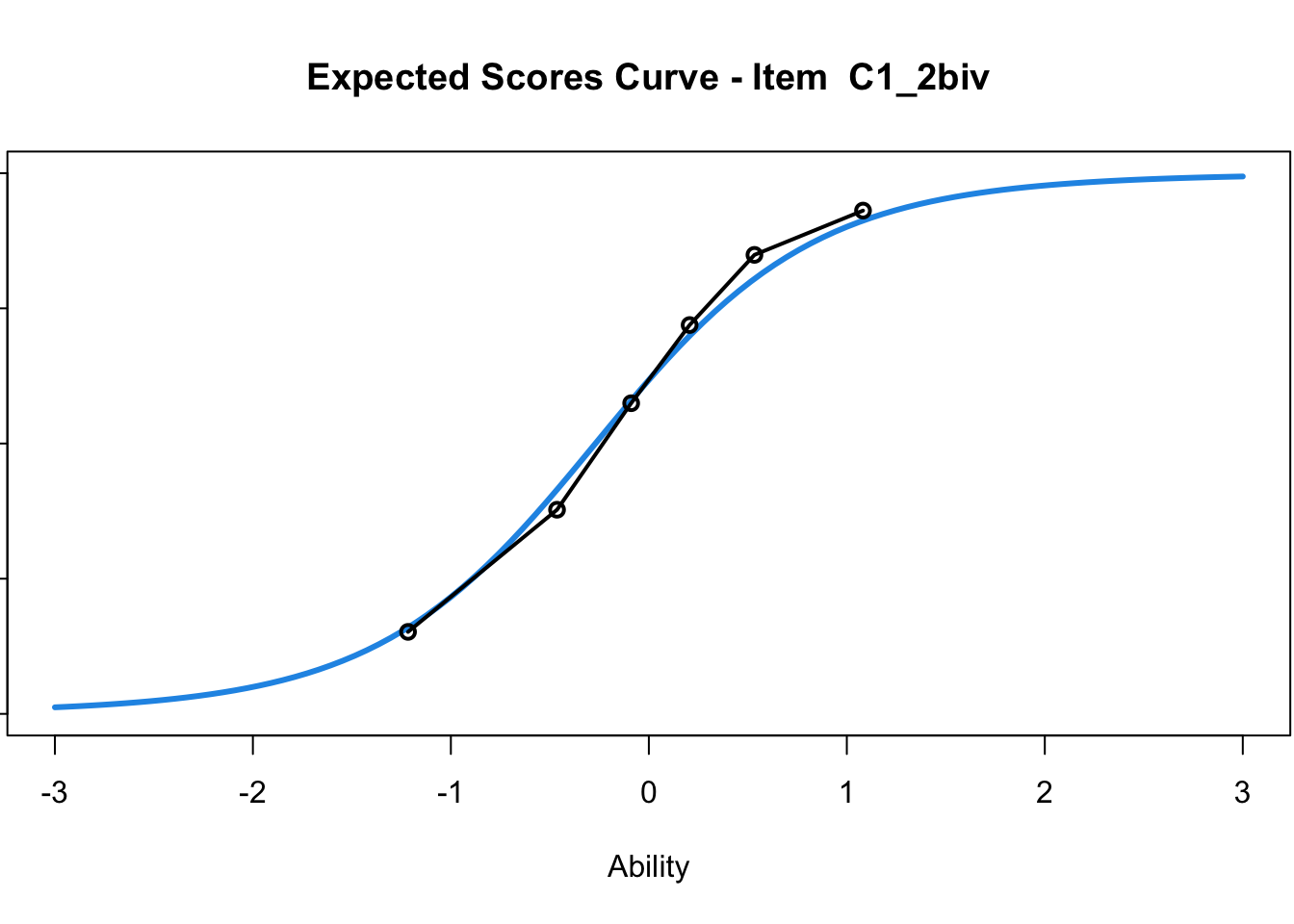
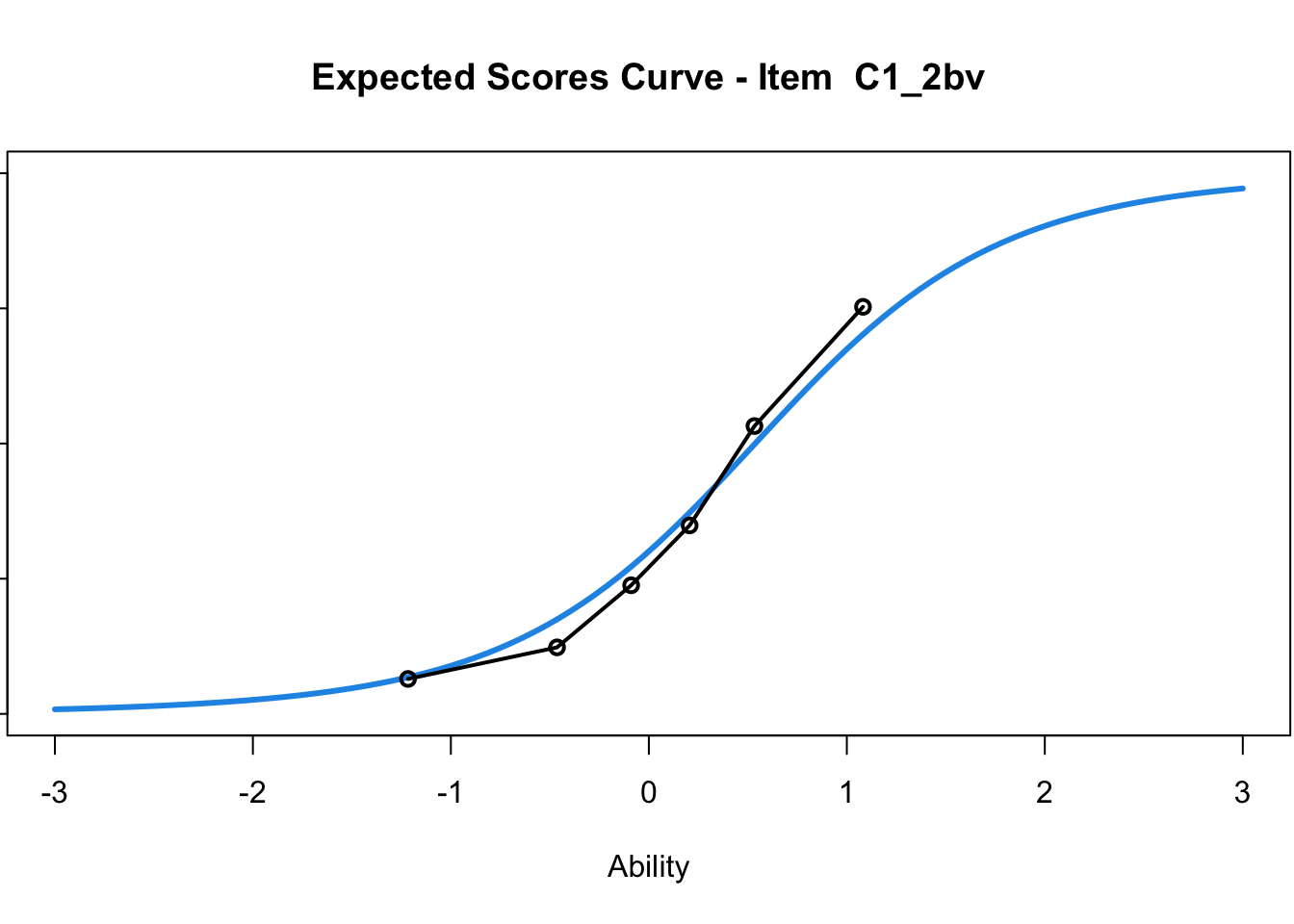
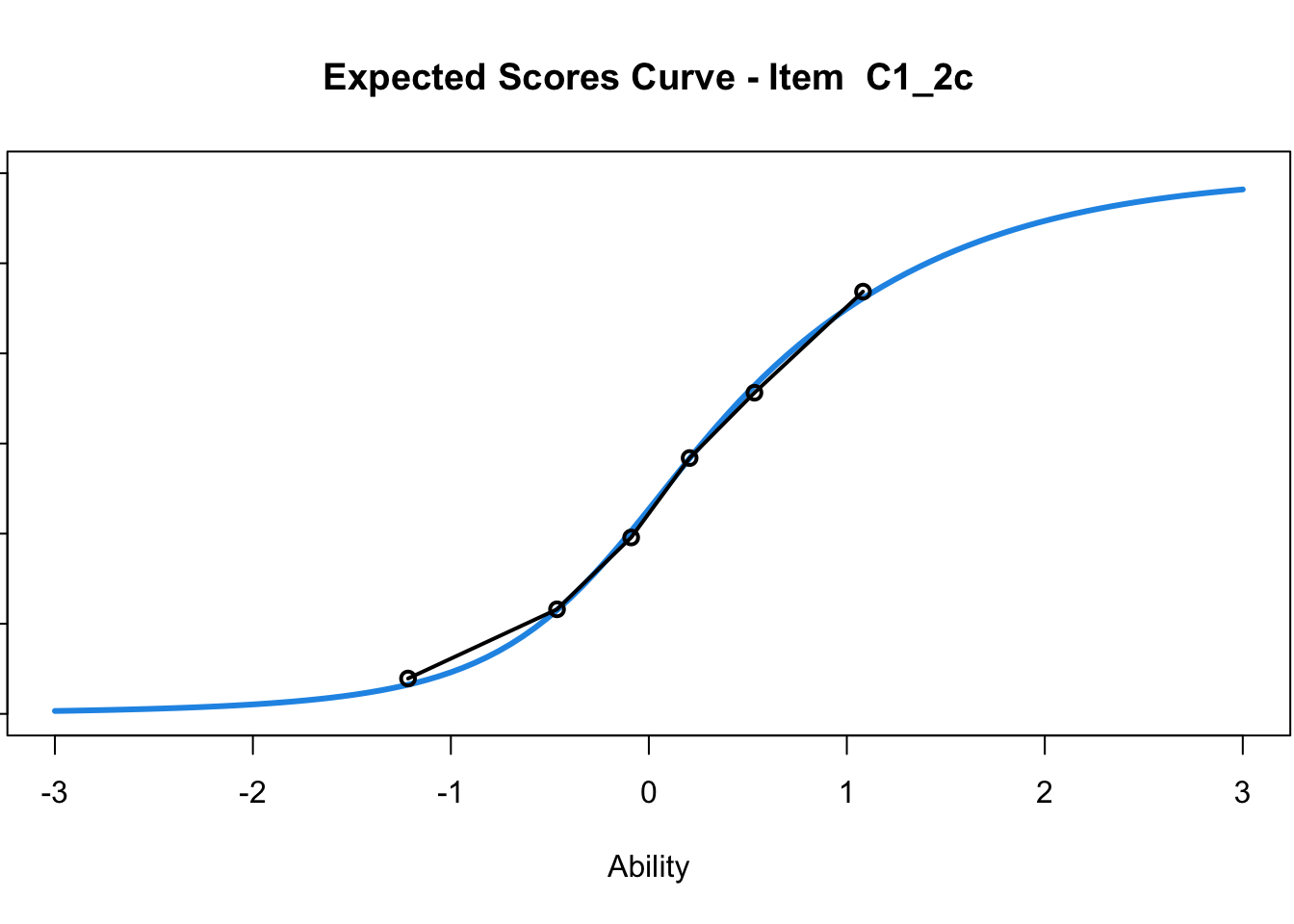
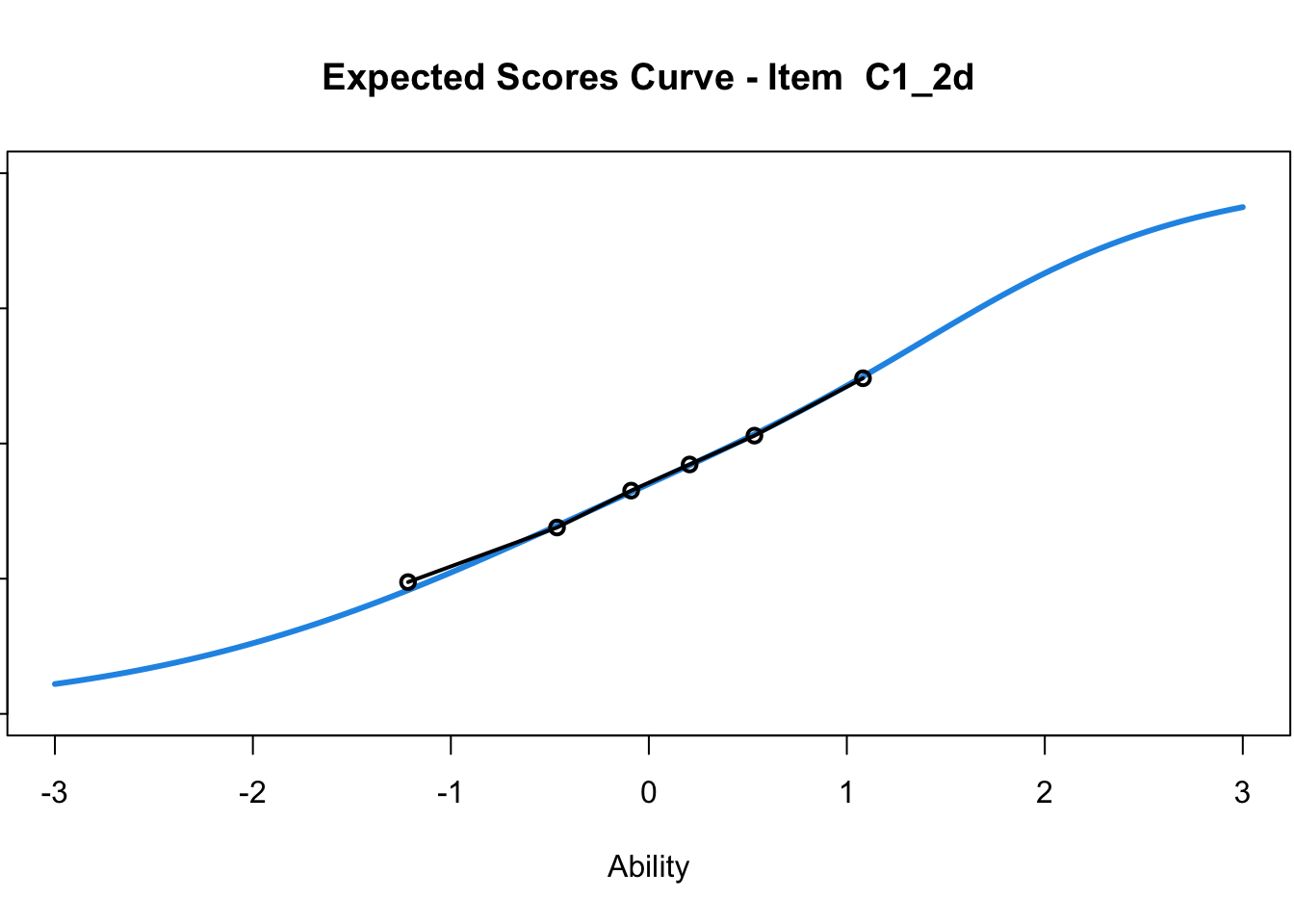
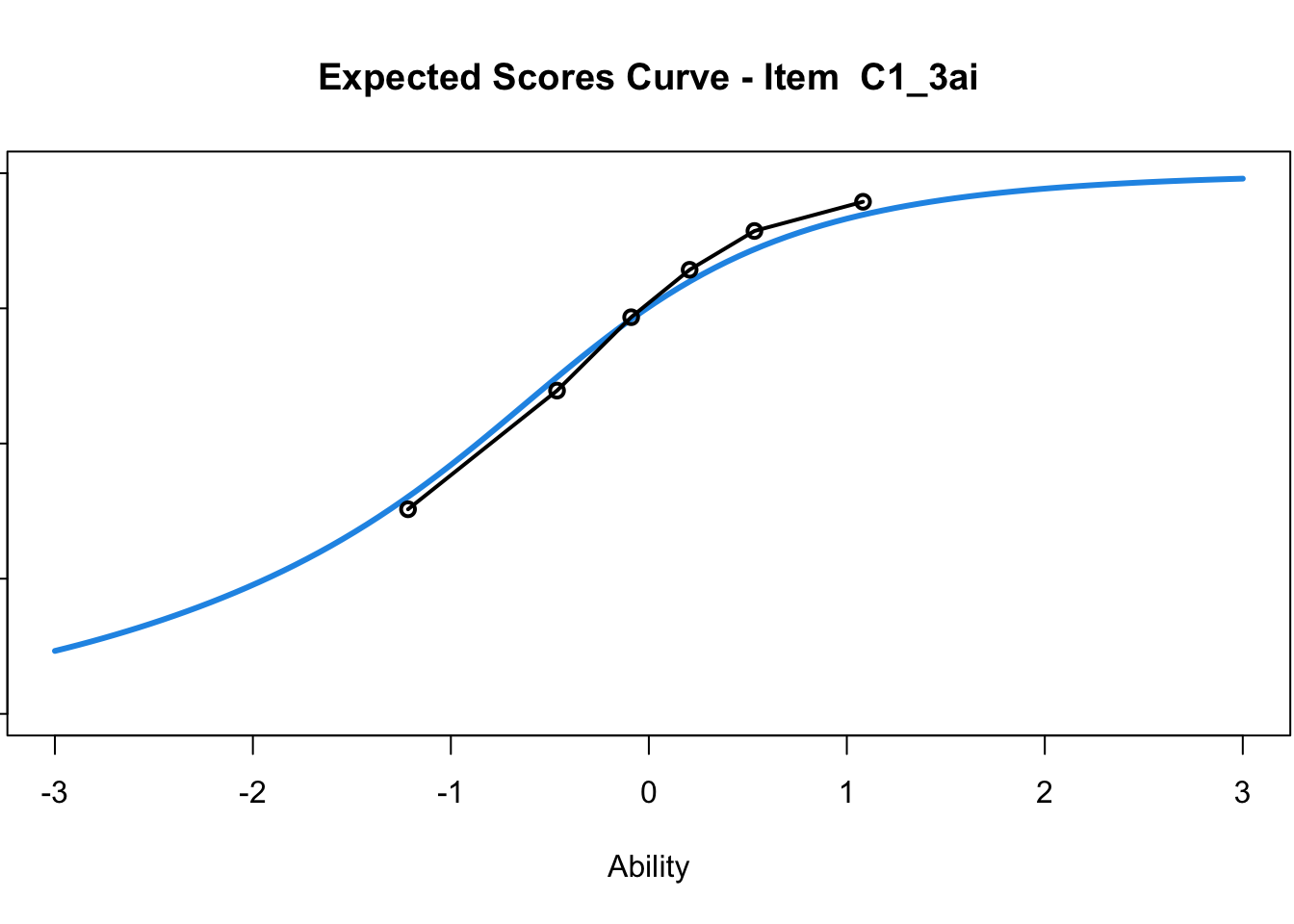
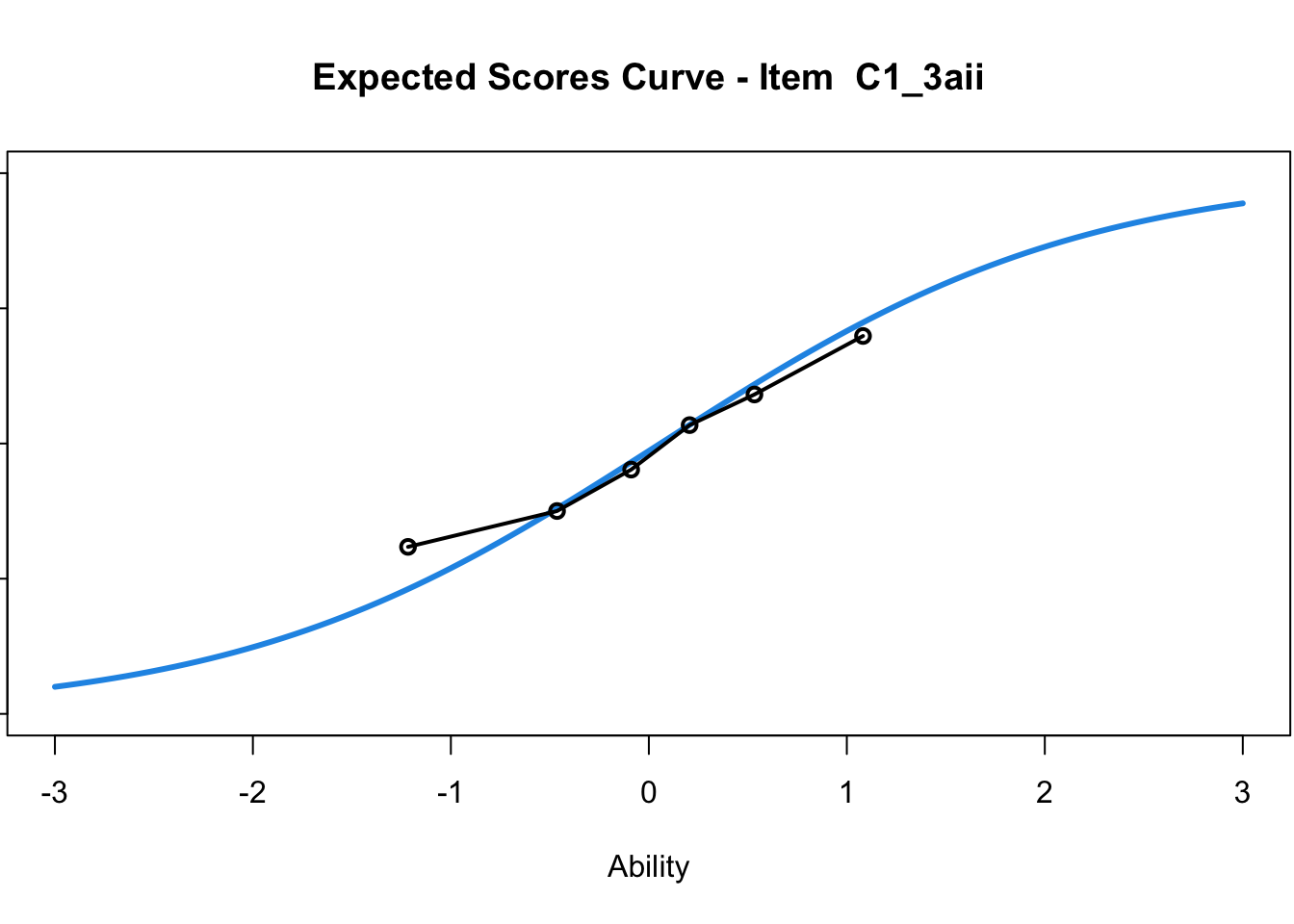
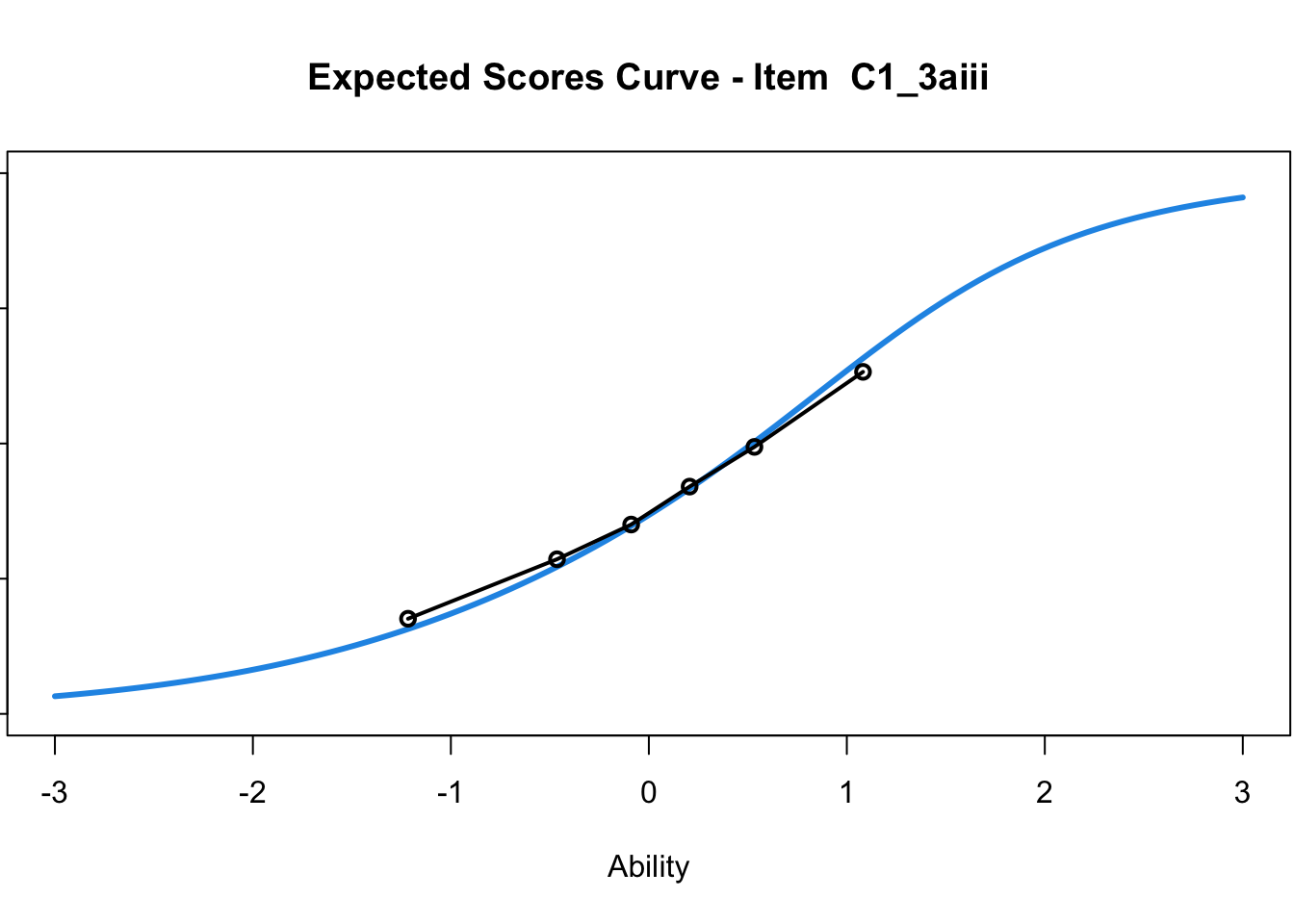
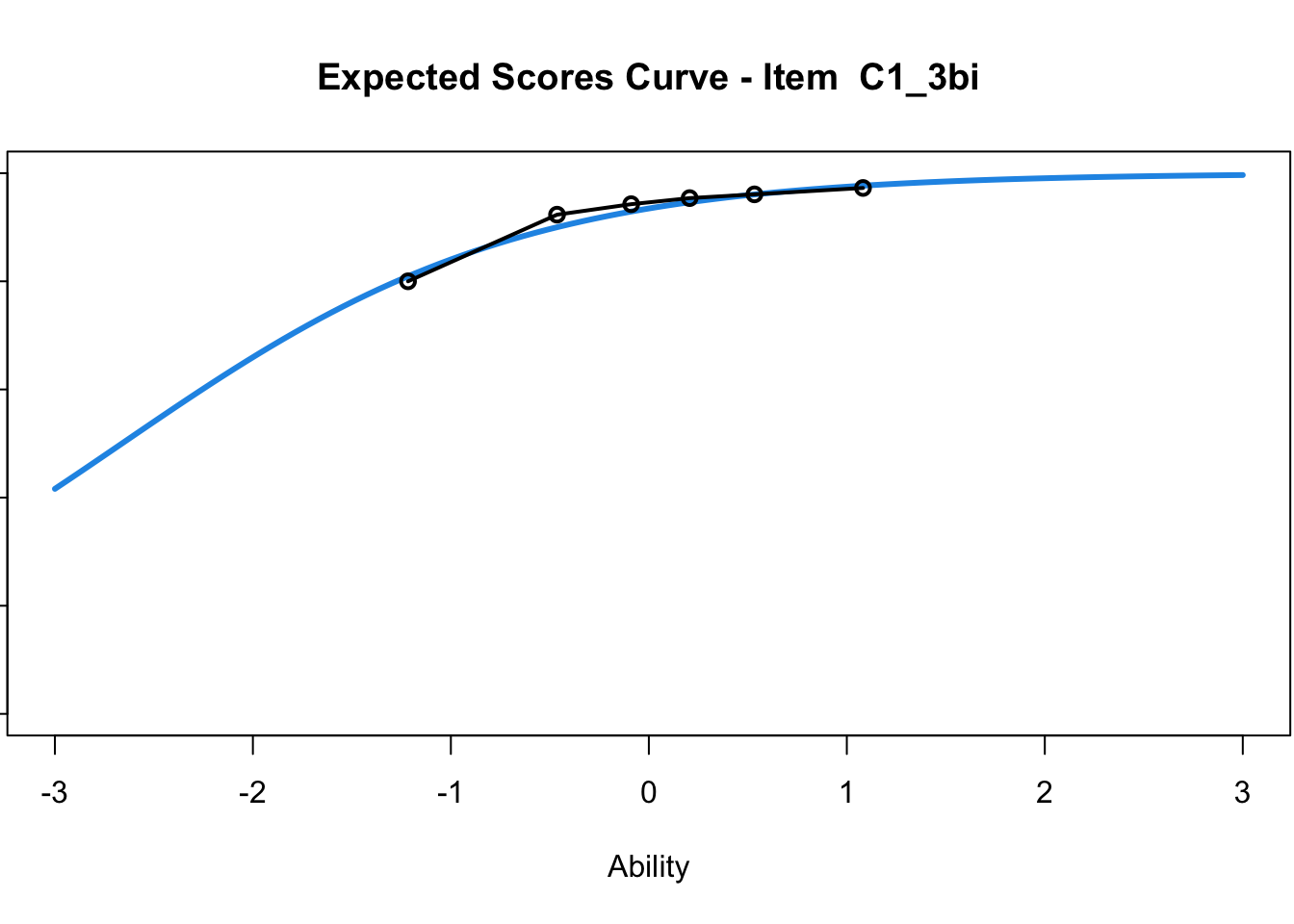
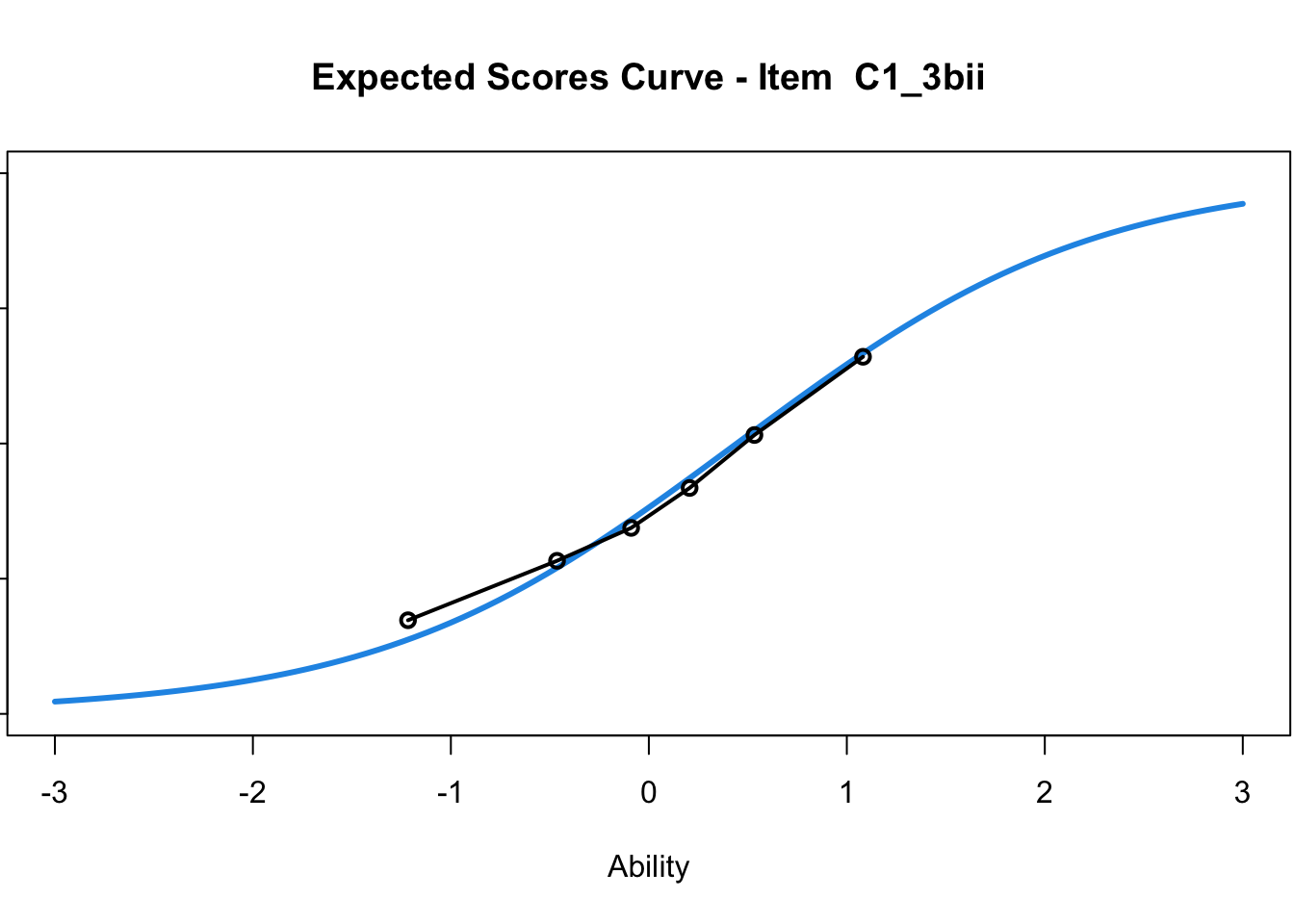
....................................................
Plots exported in png format into folder:
/Users/chris/Documents/CM3/Plotsplot(mod1,type="items") #ICCs
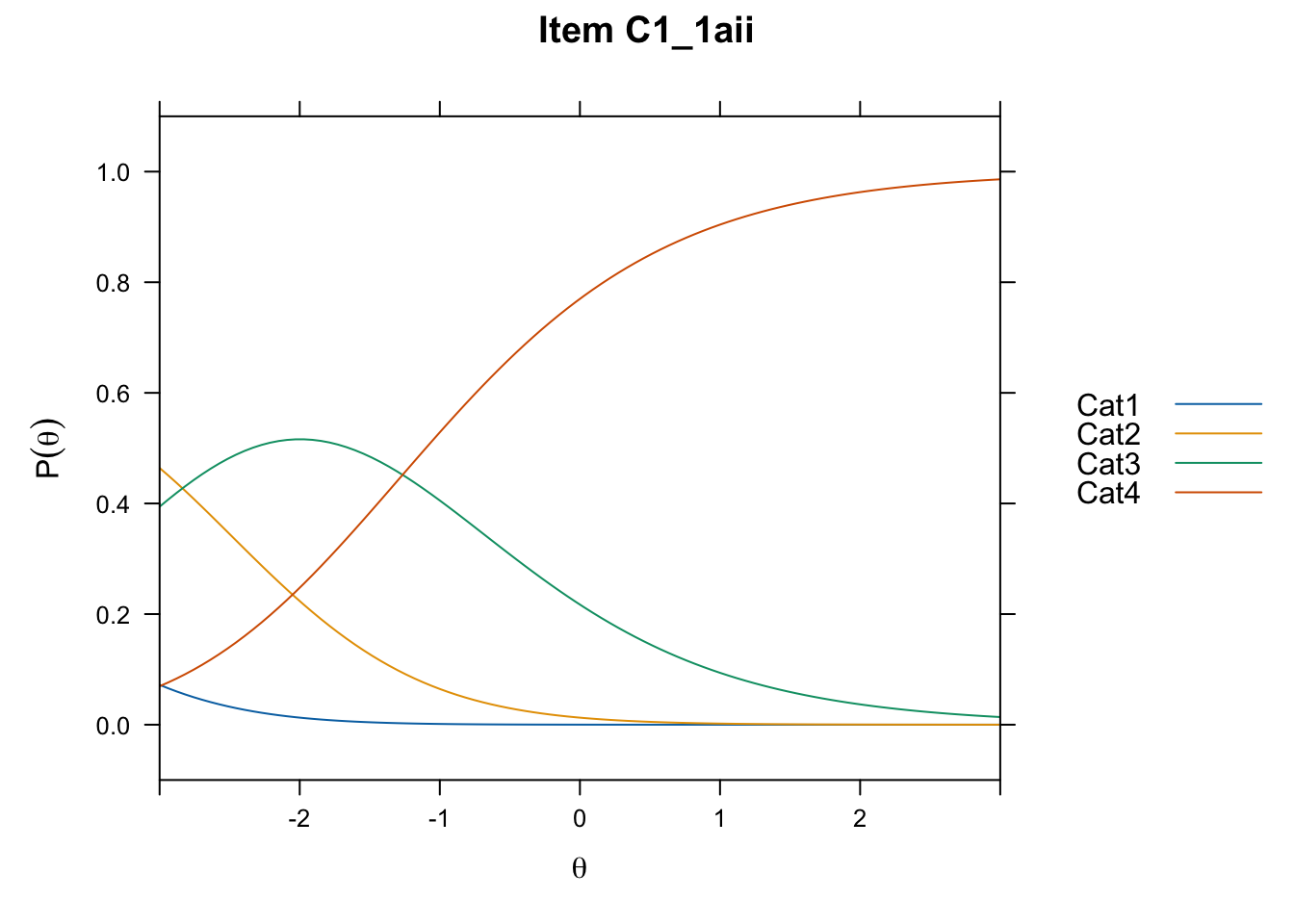
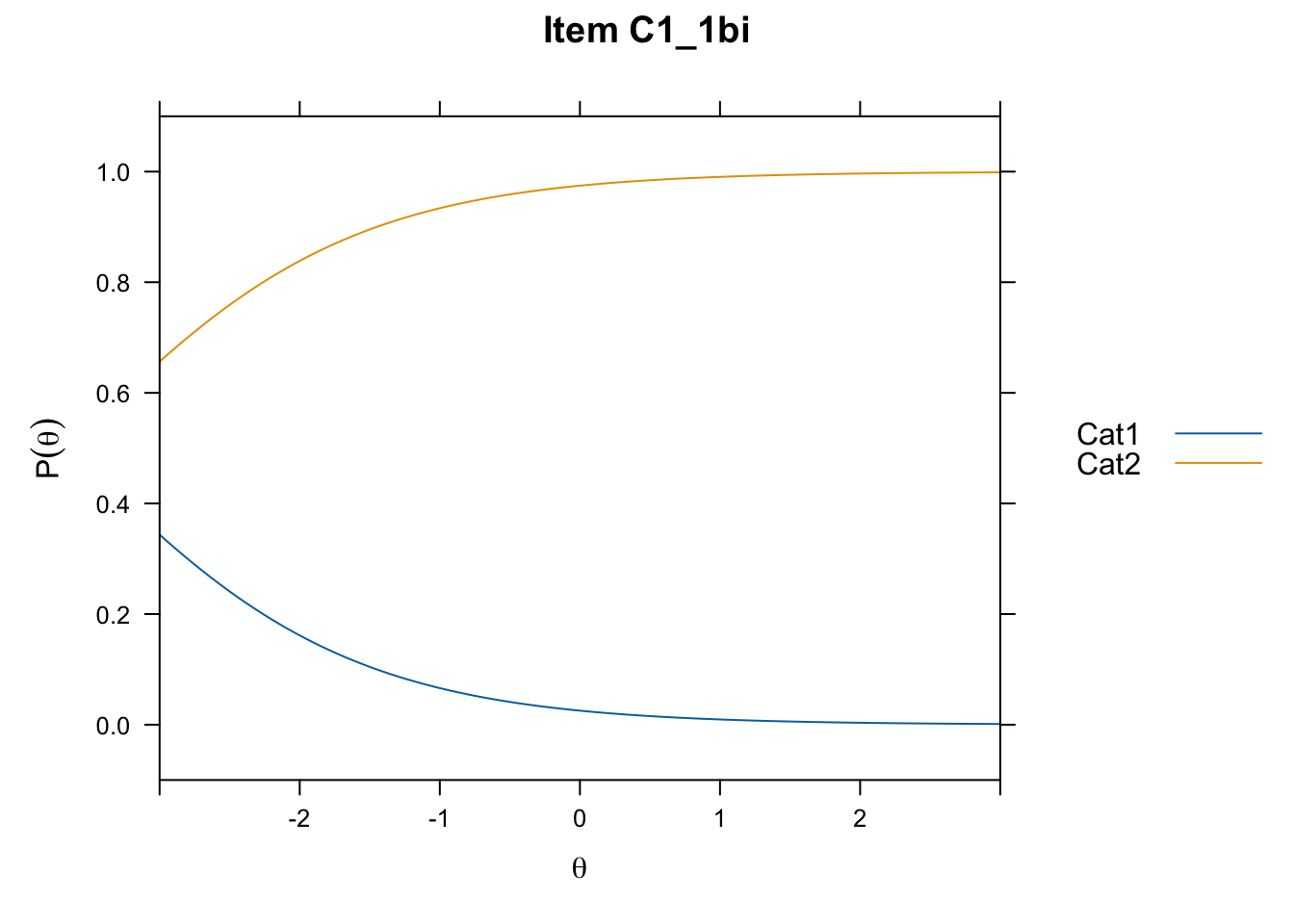
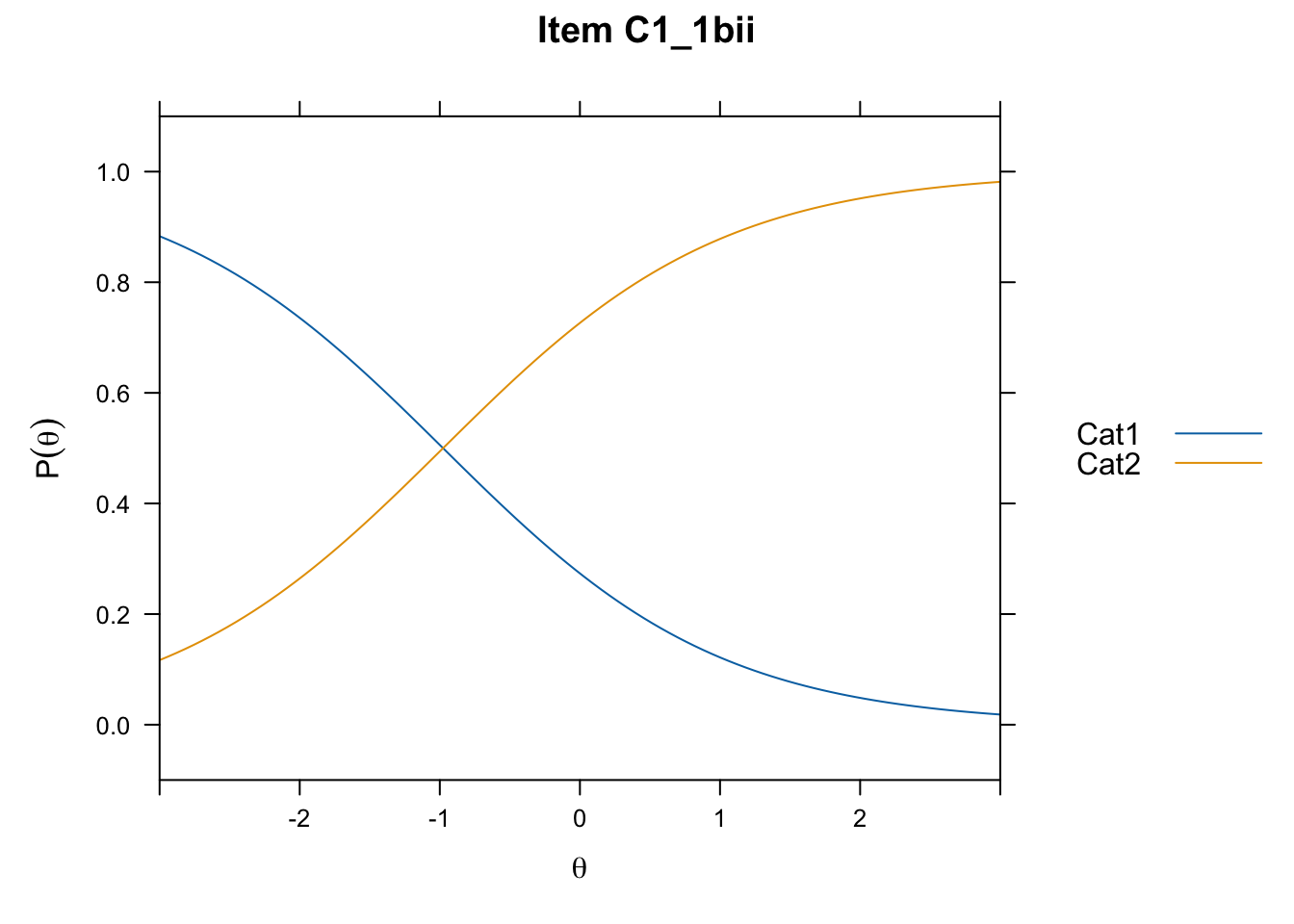
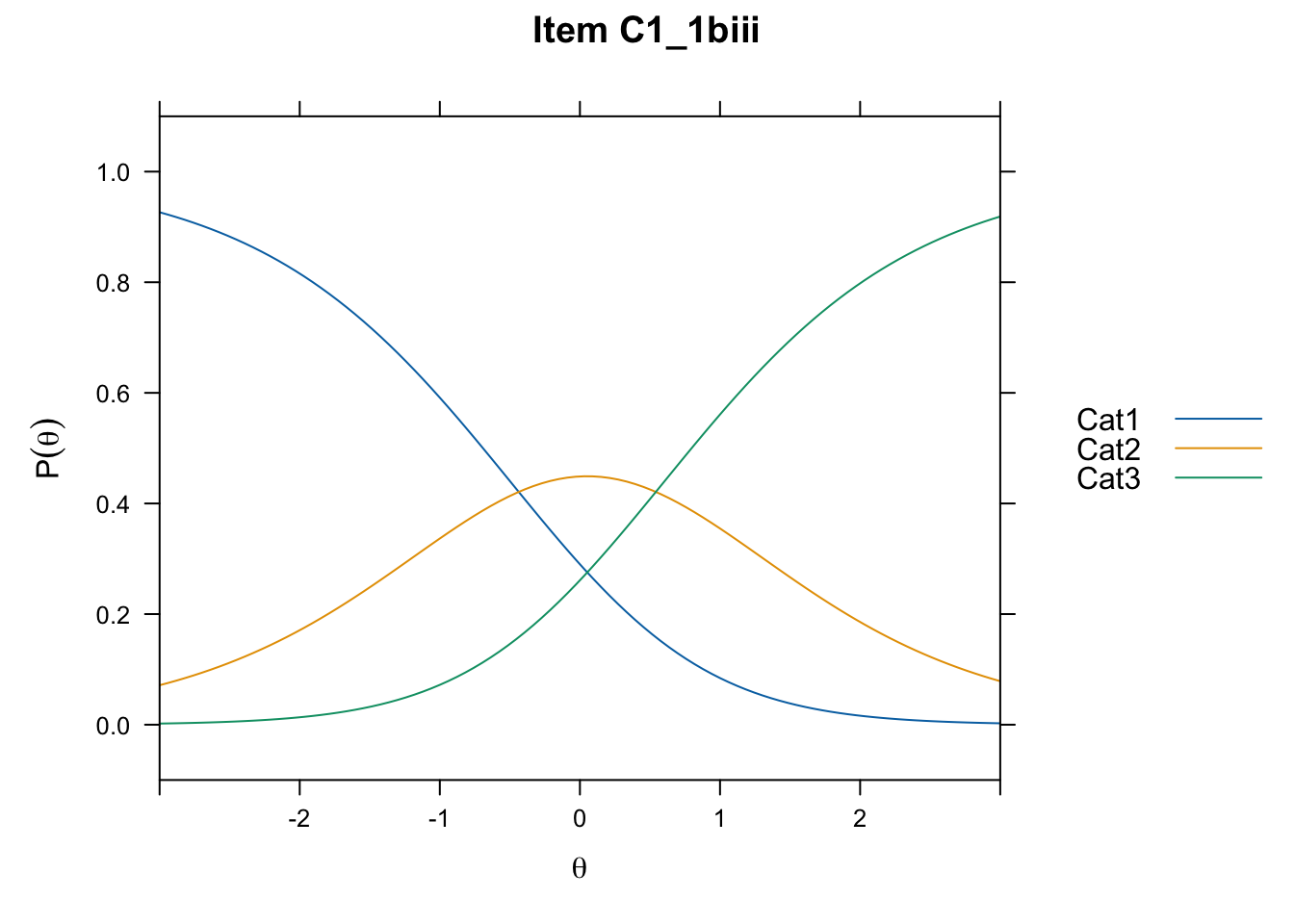
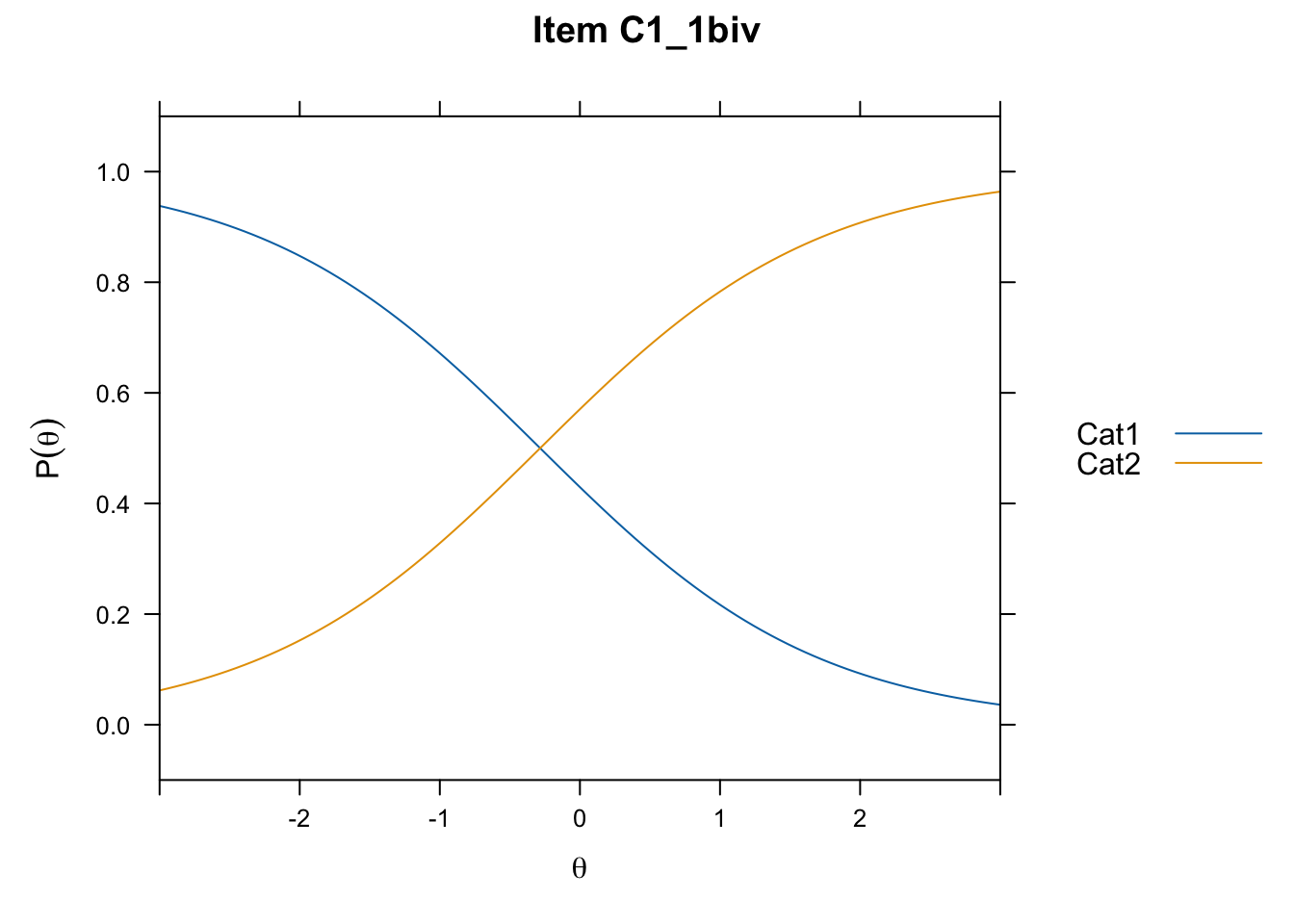
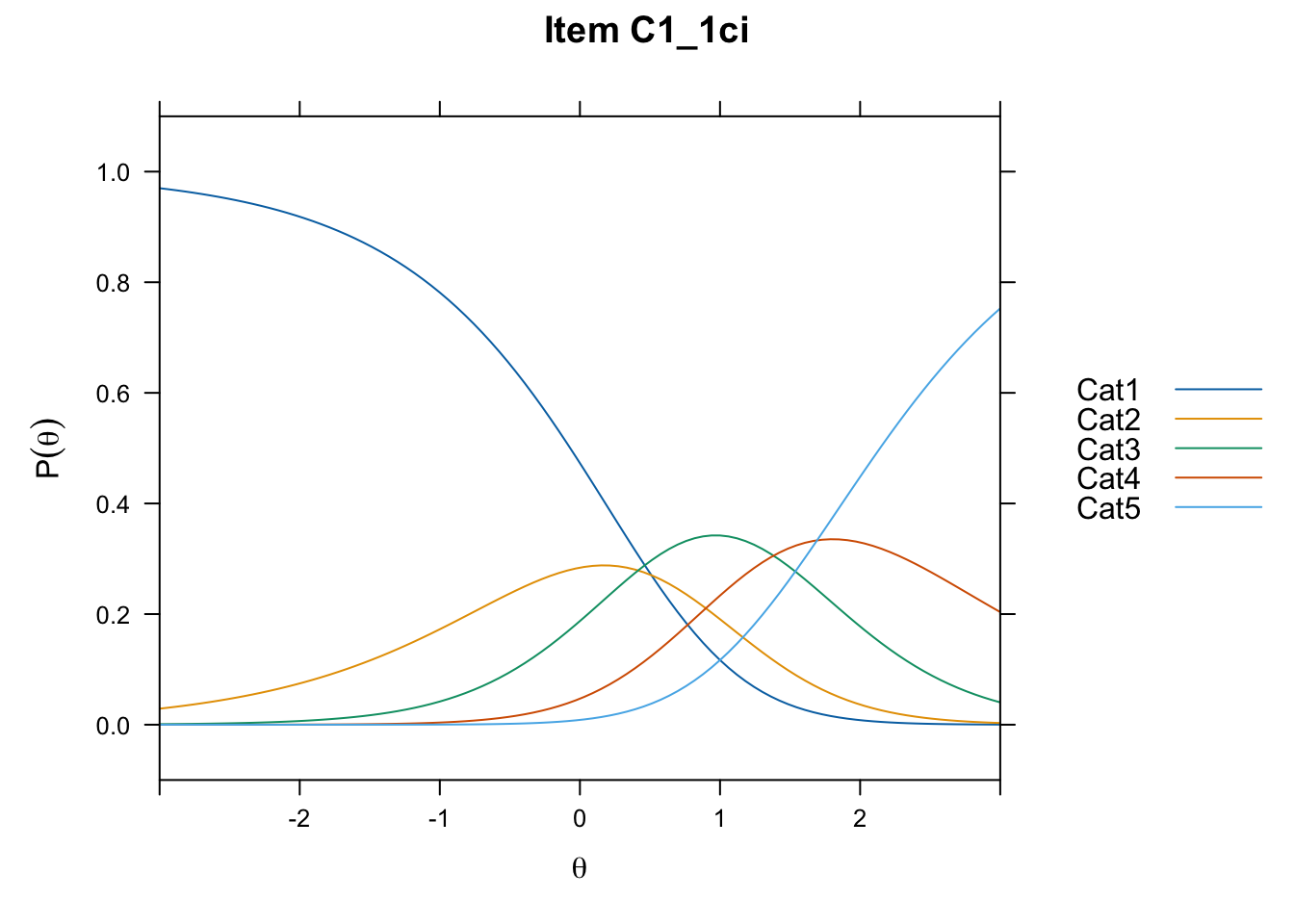
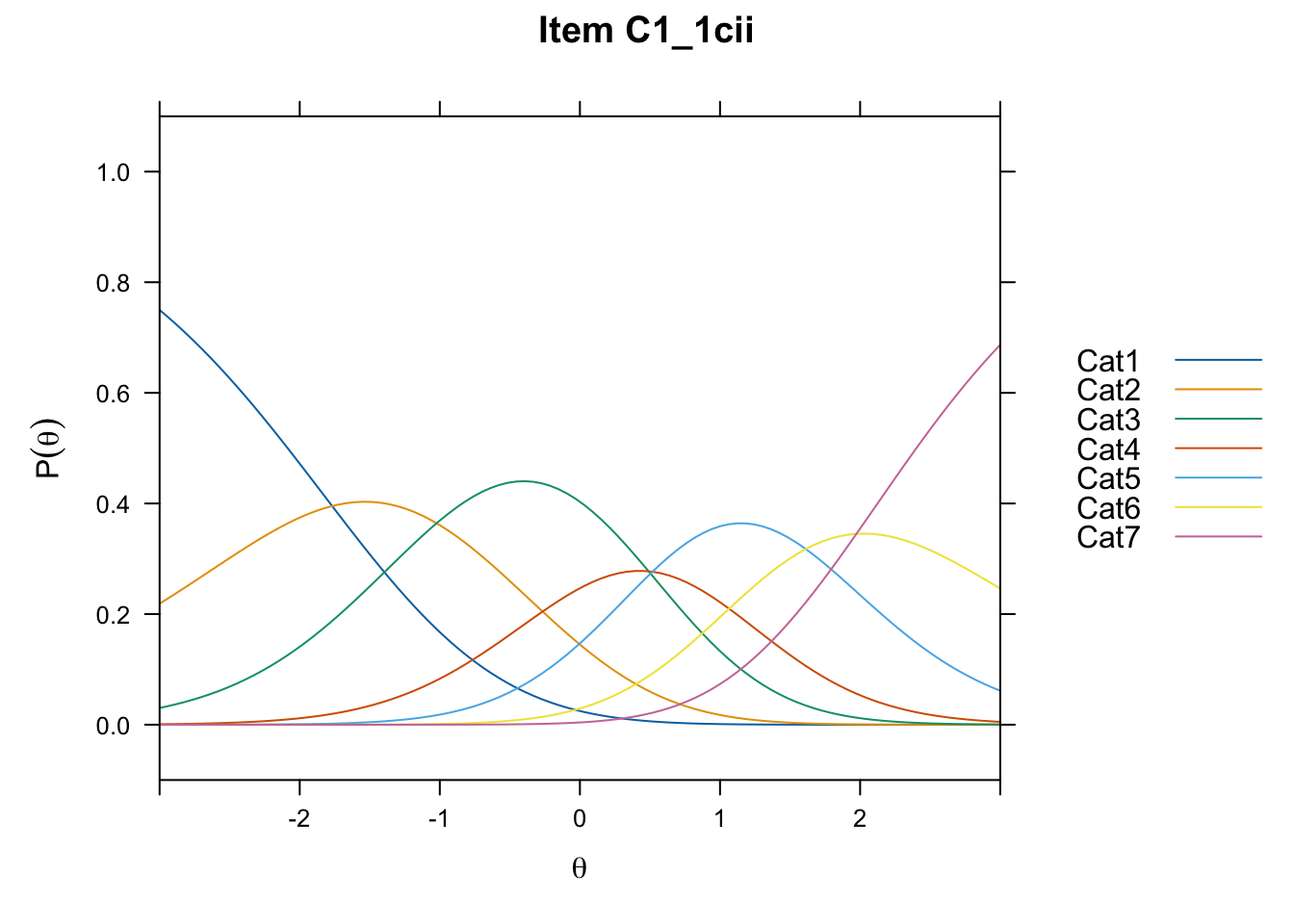
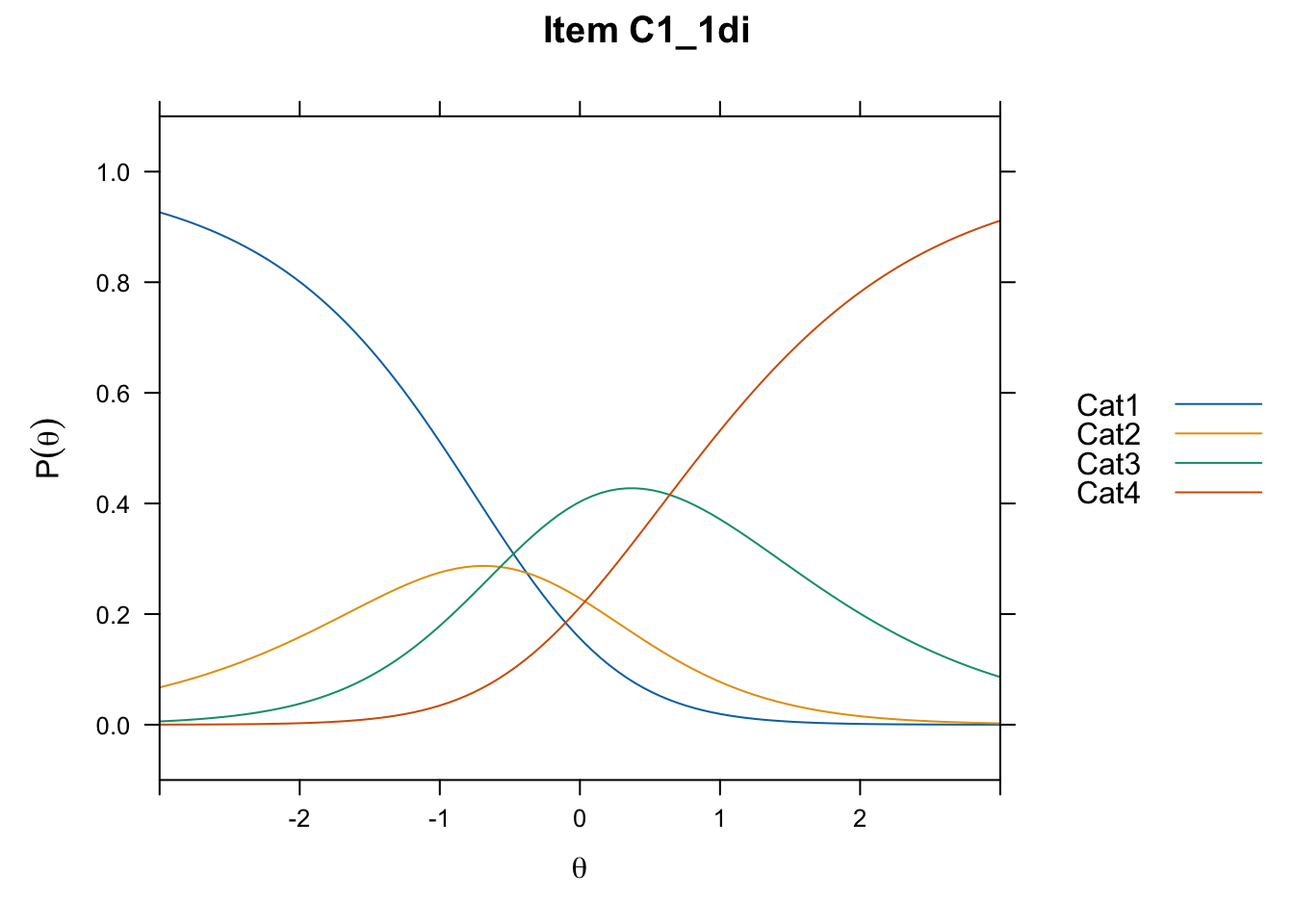
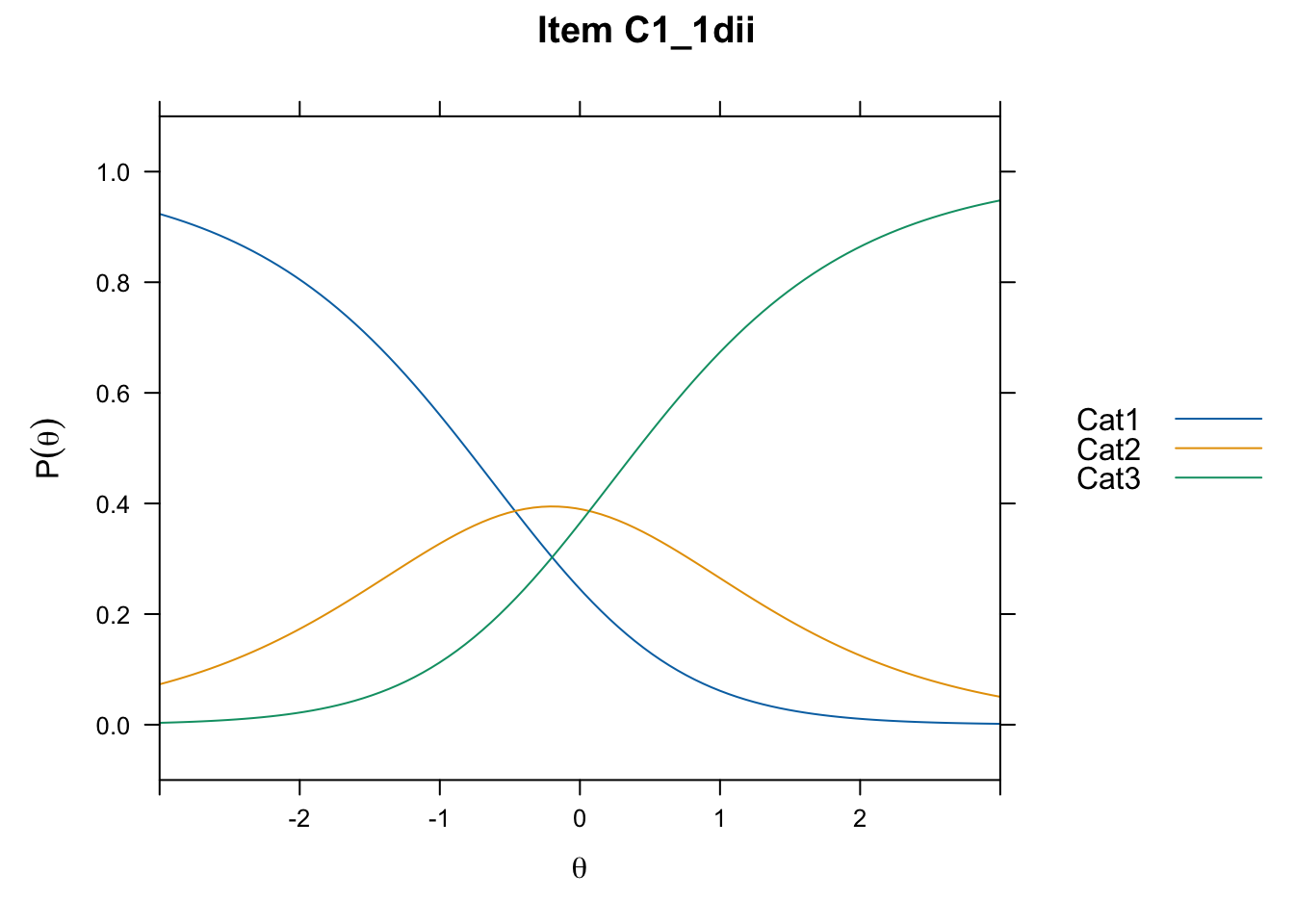
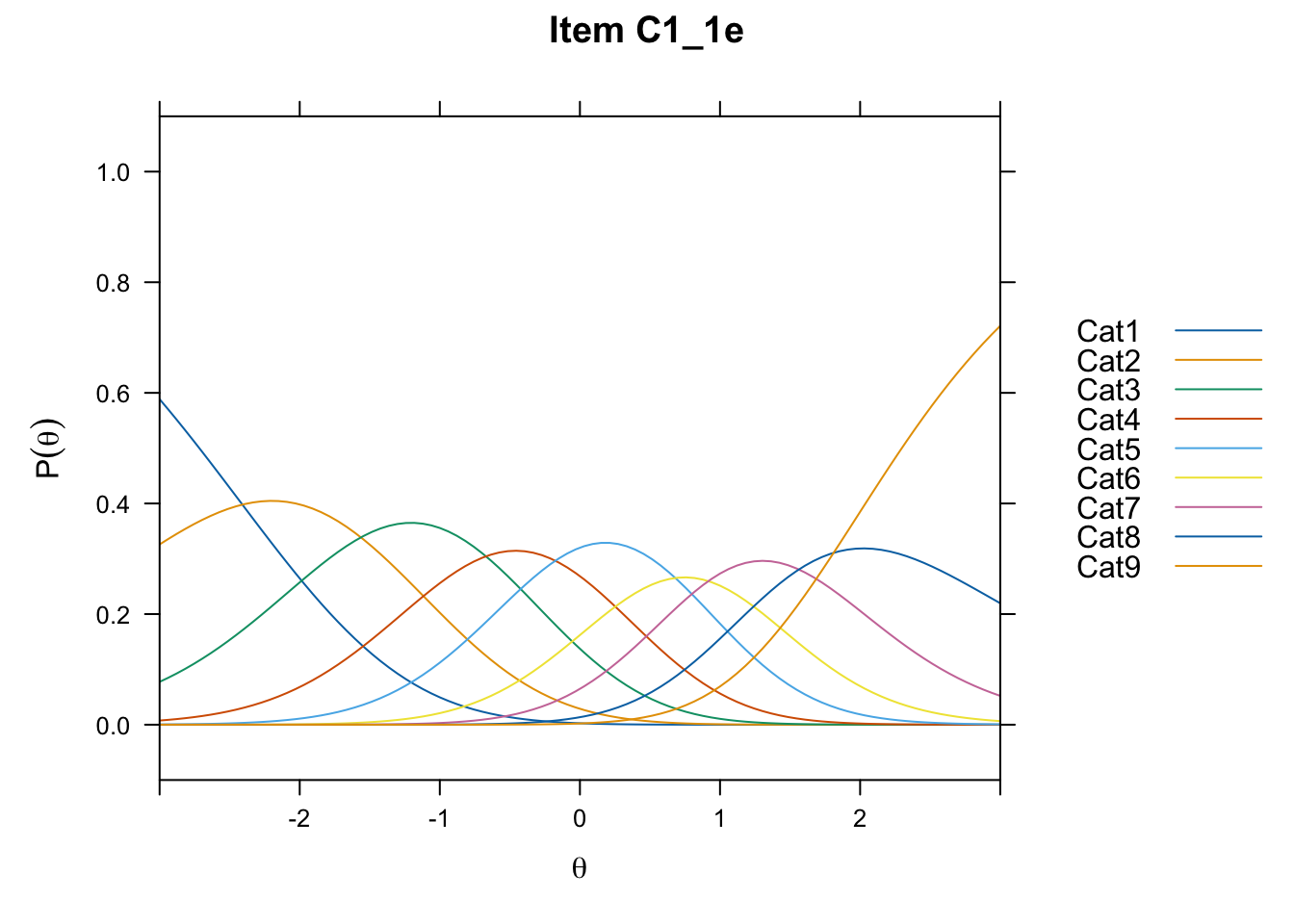
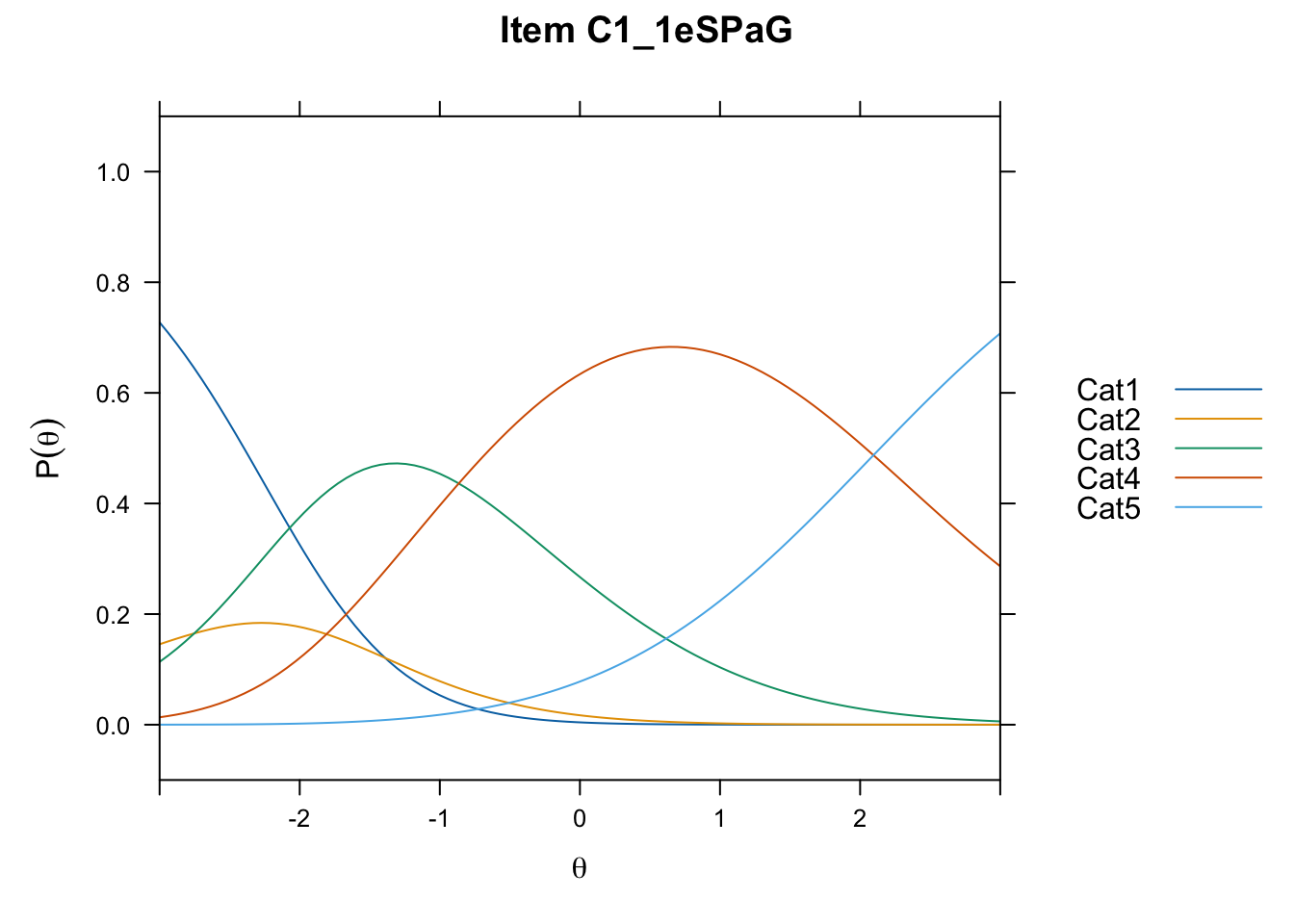
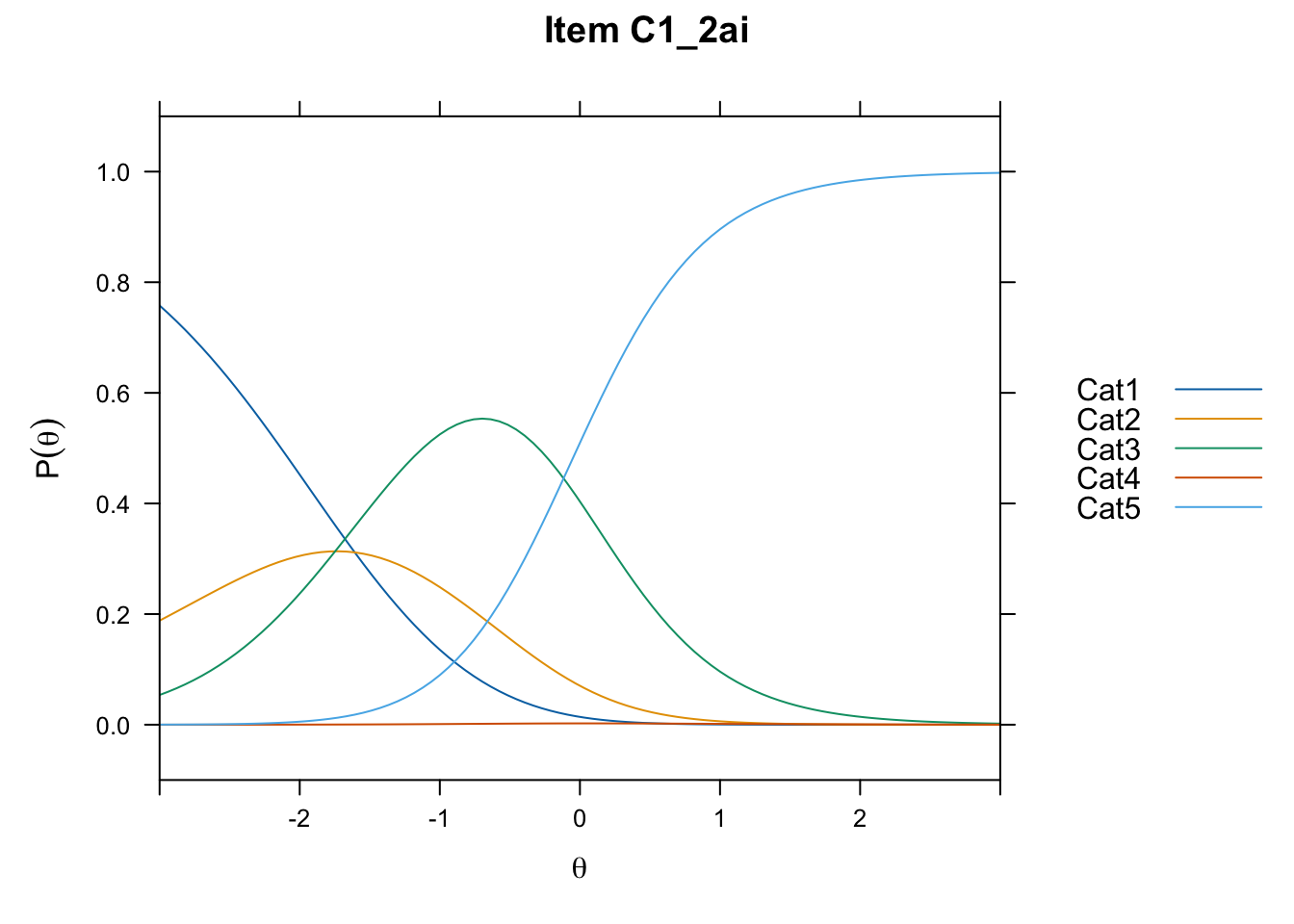
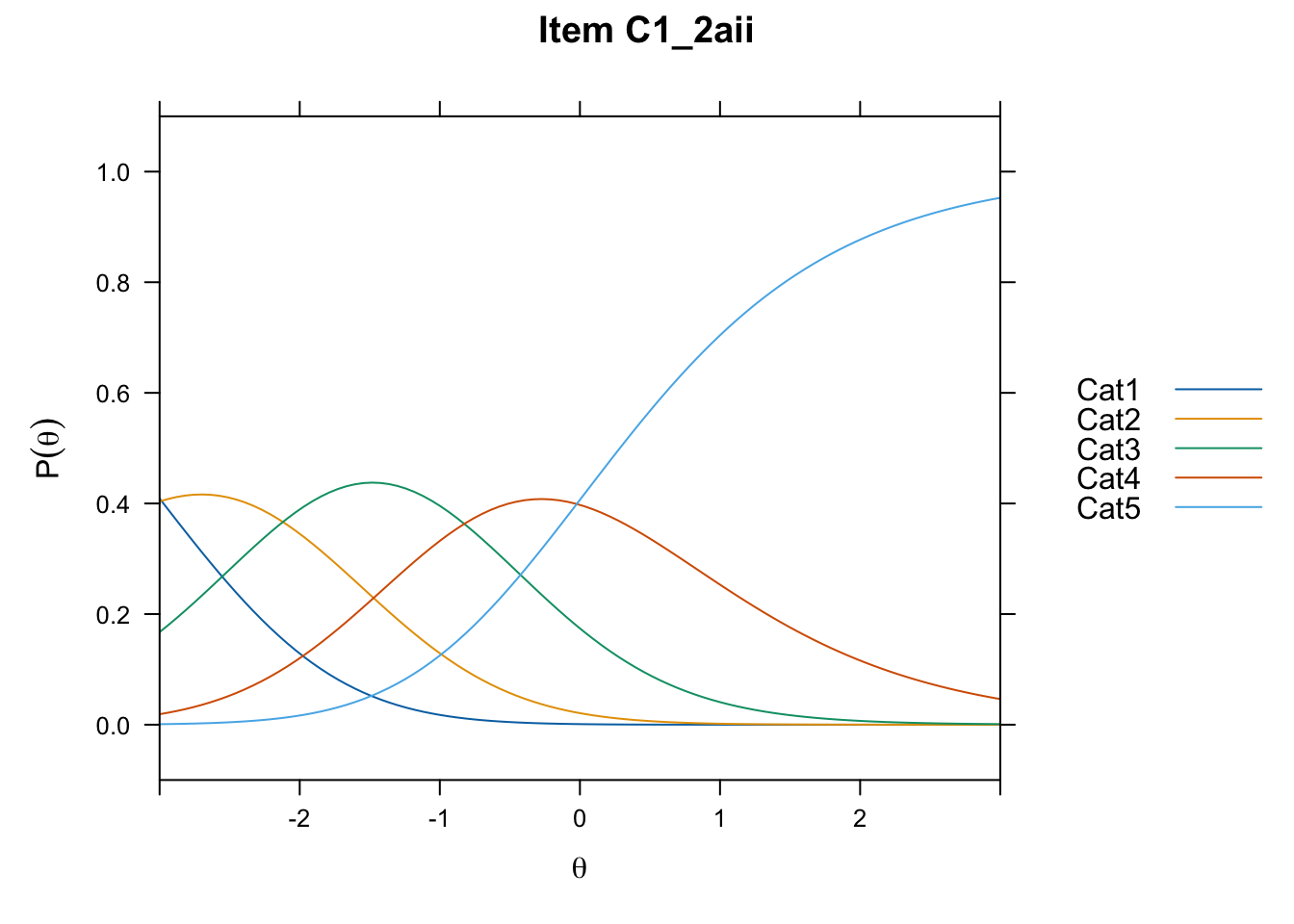
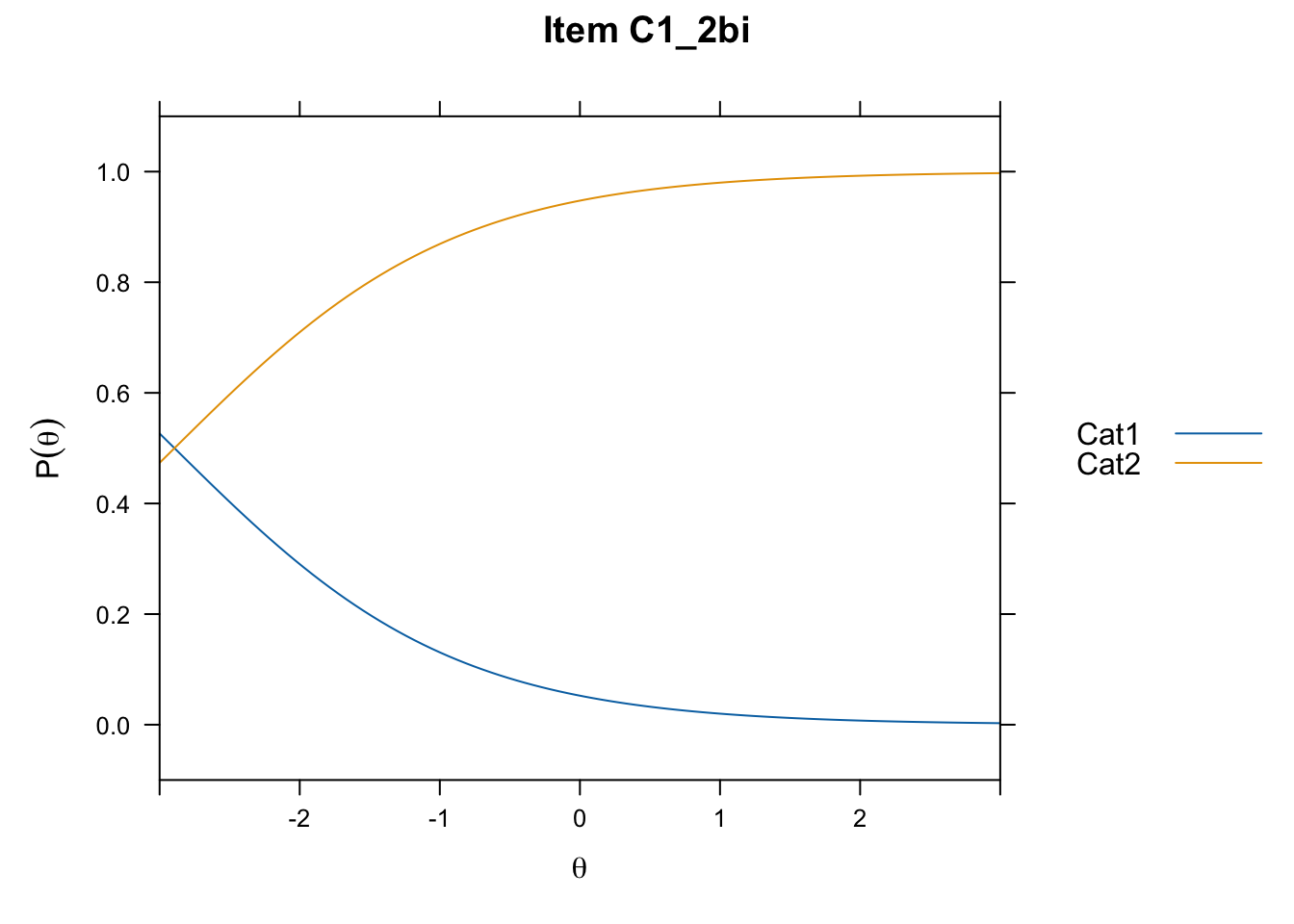
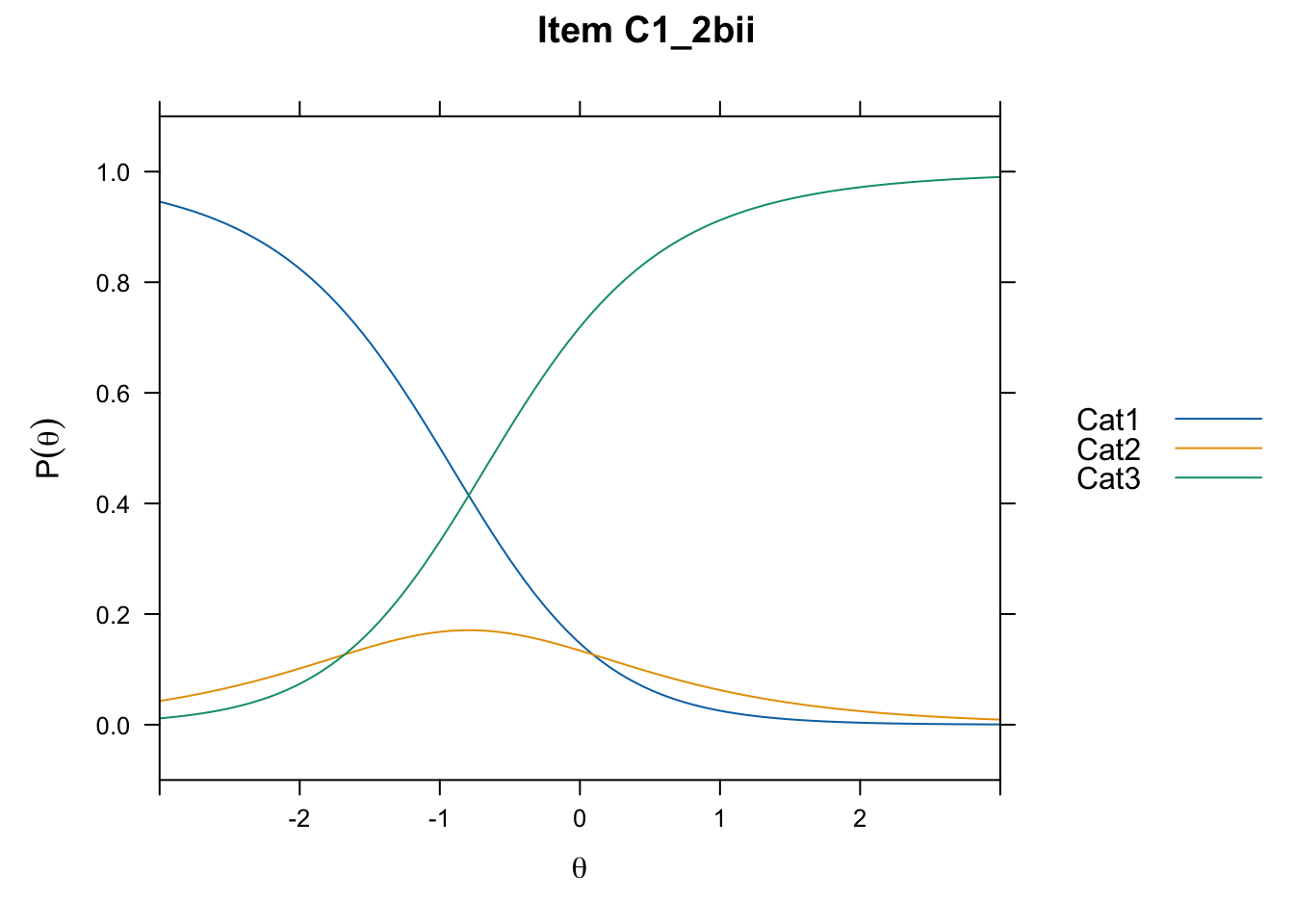
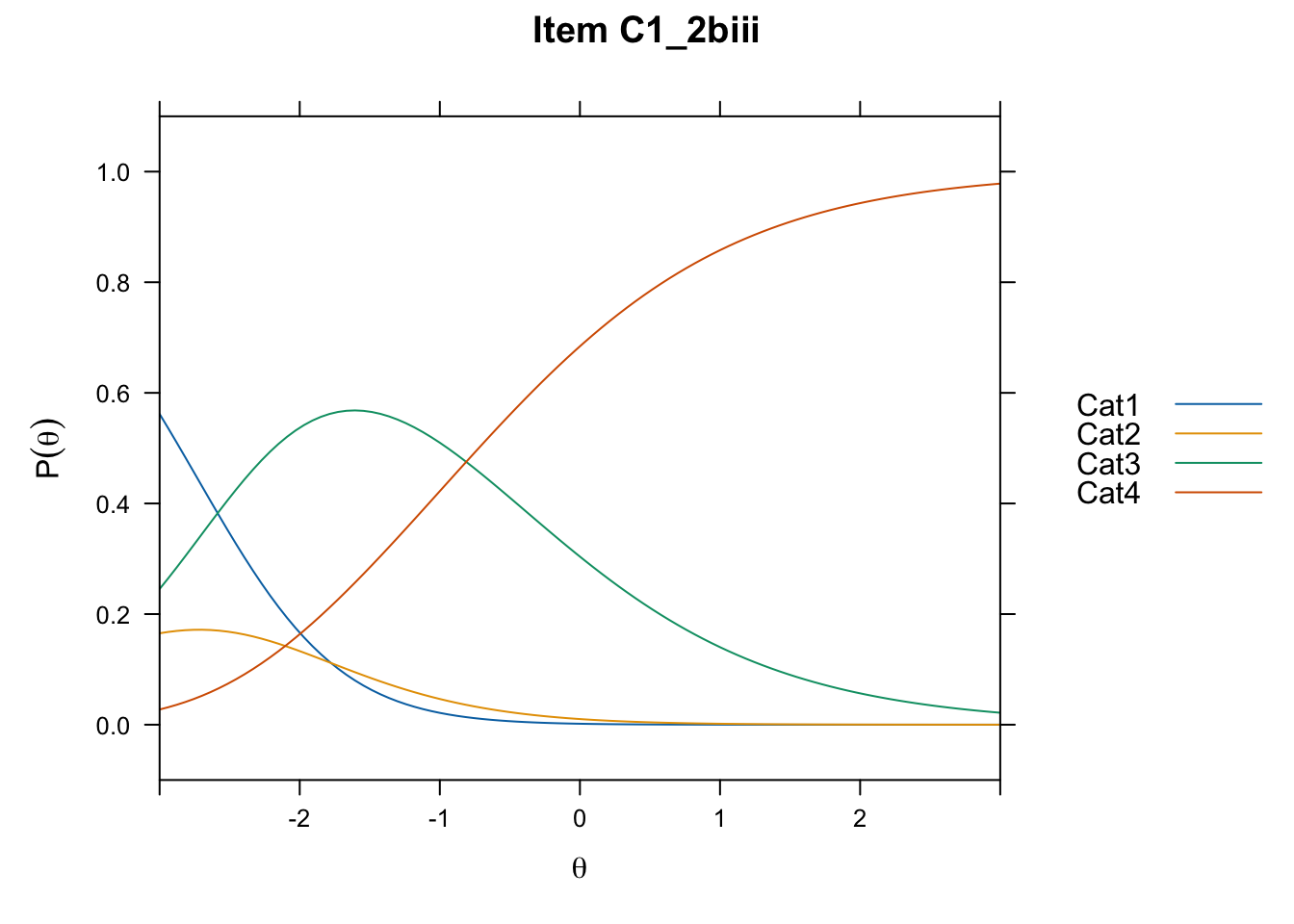
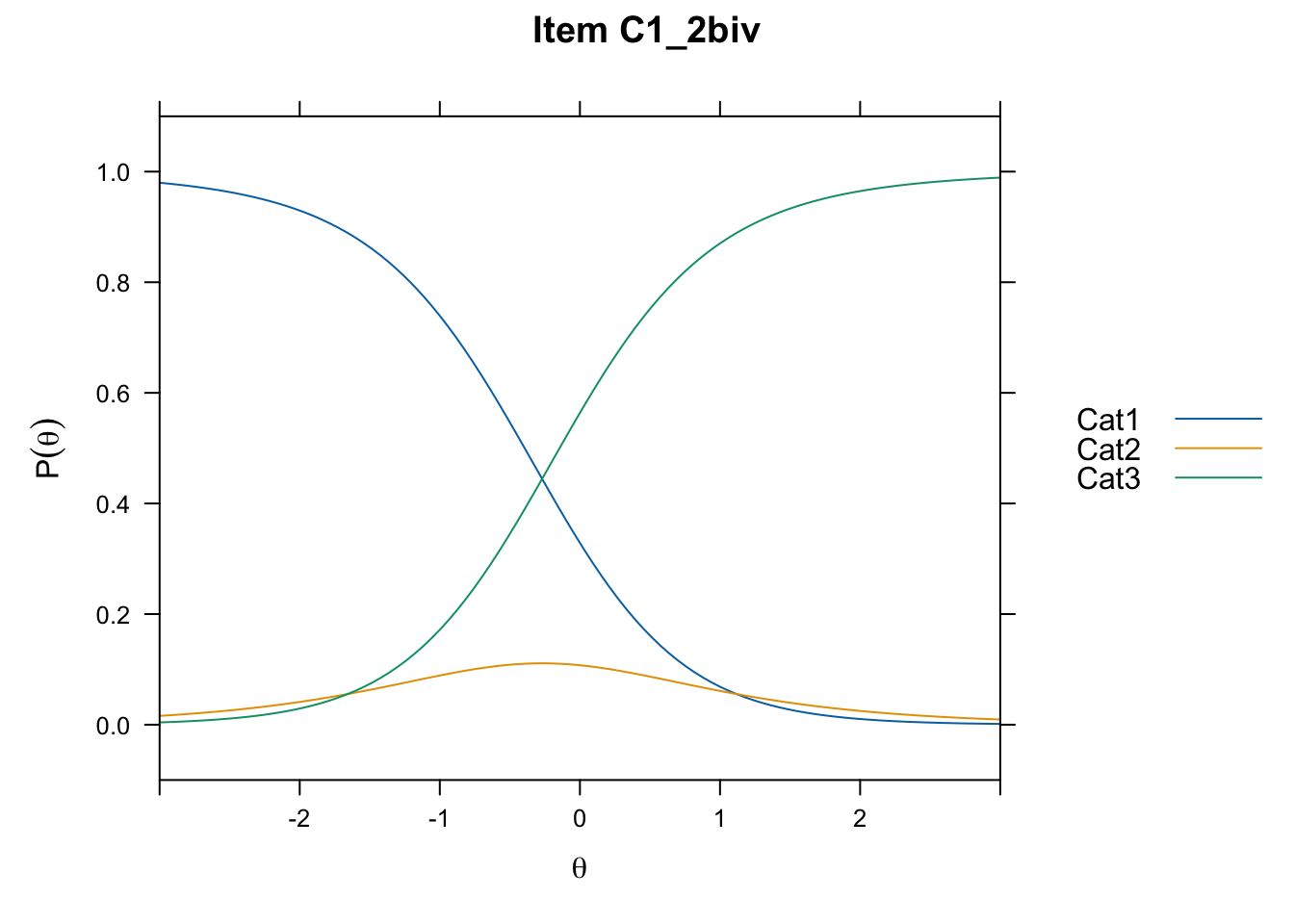
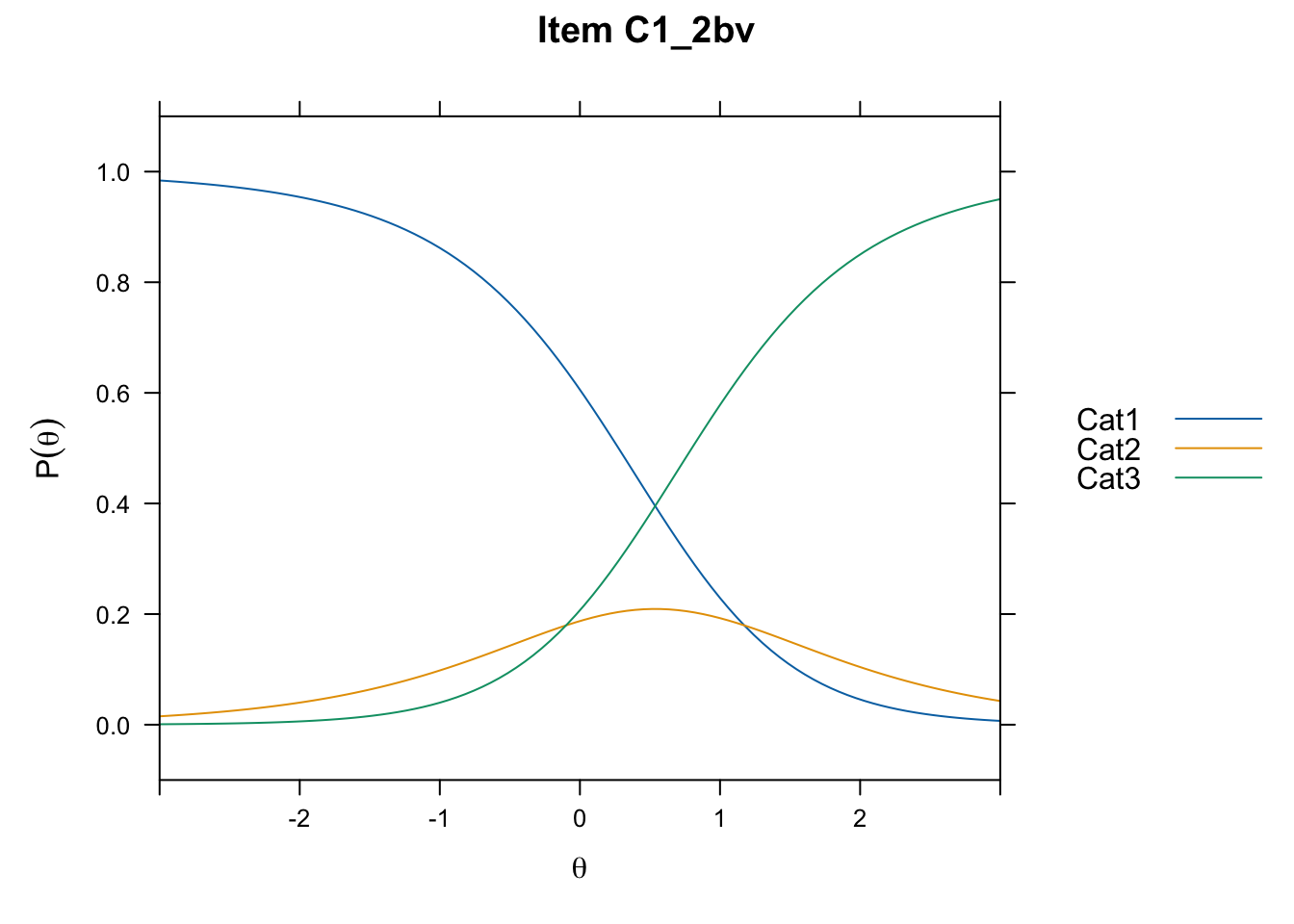
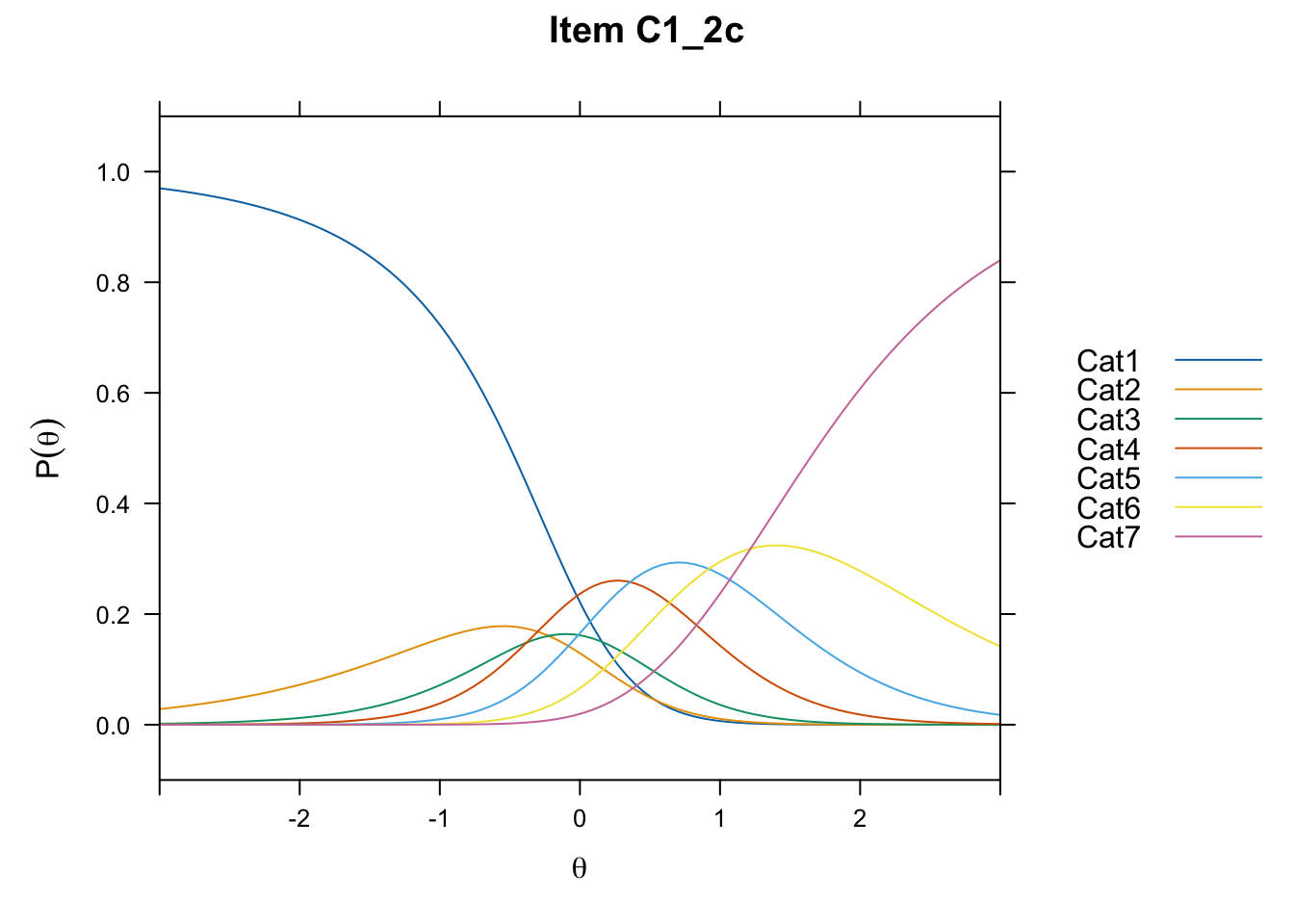
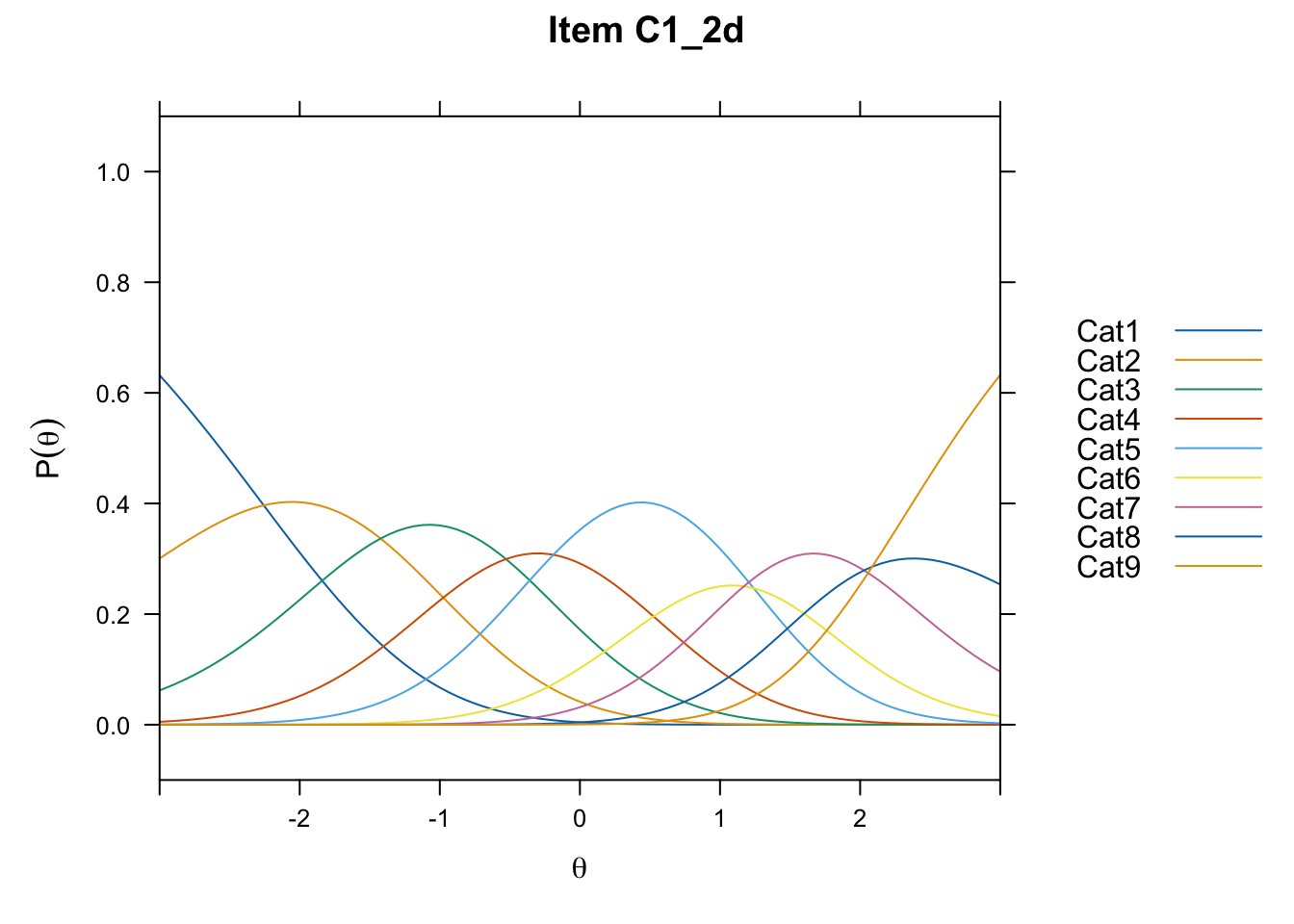
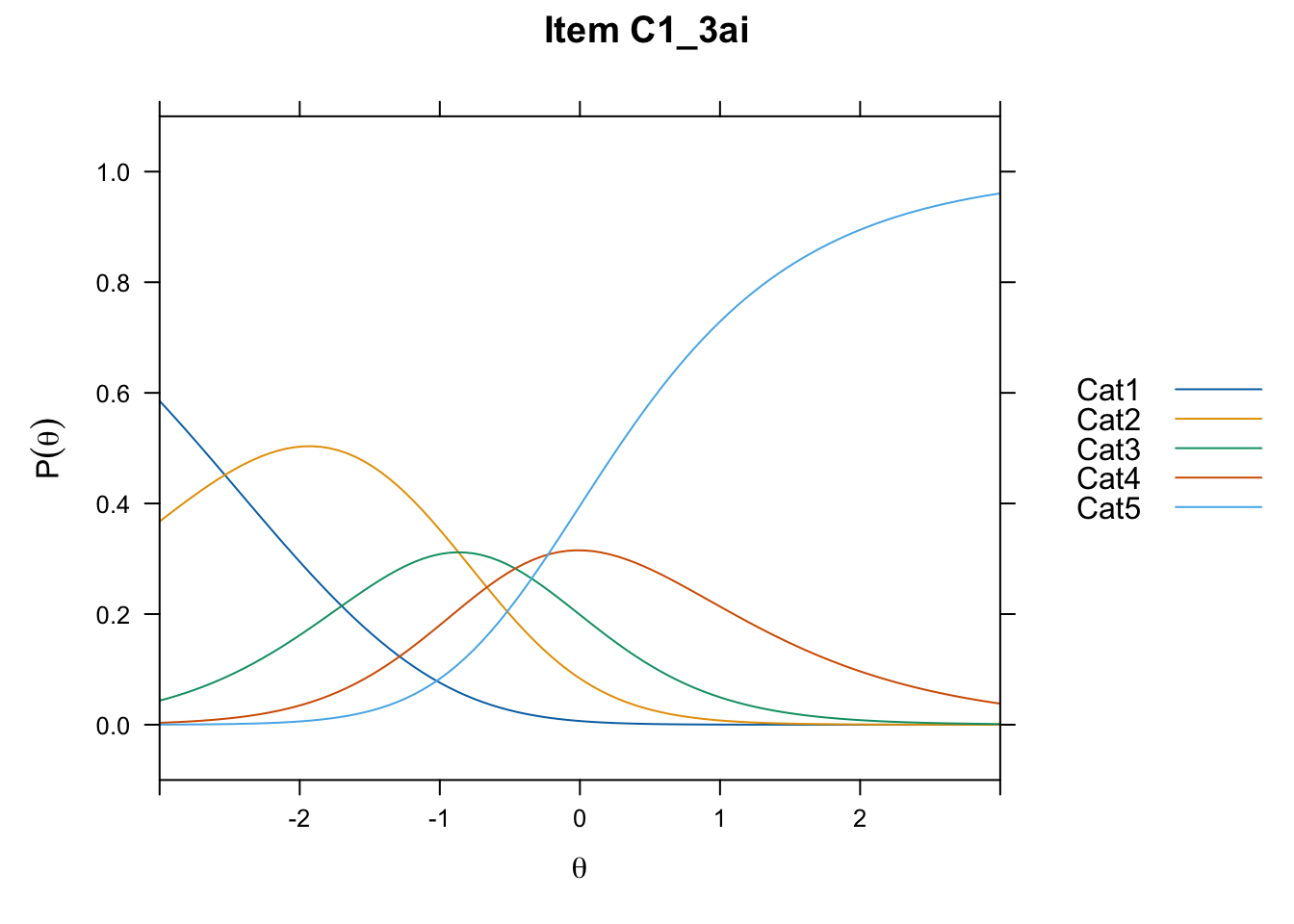
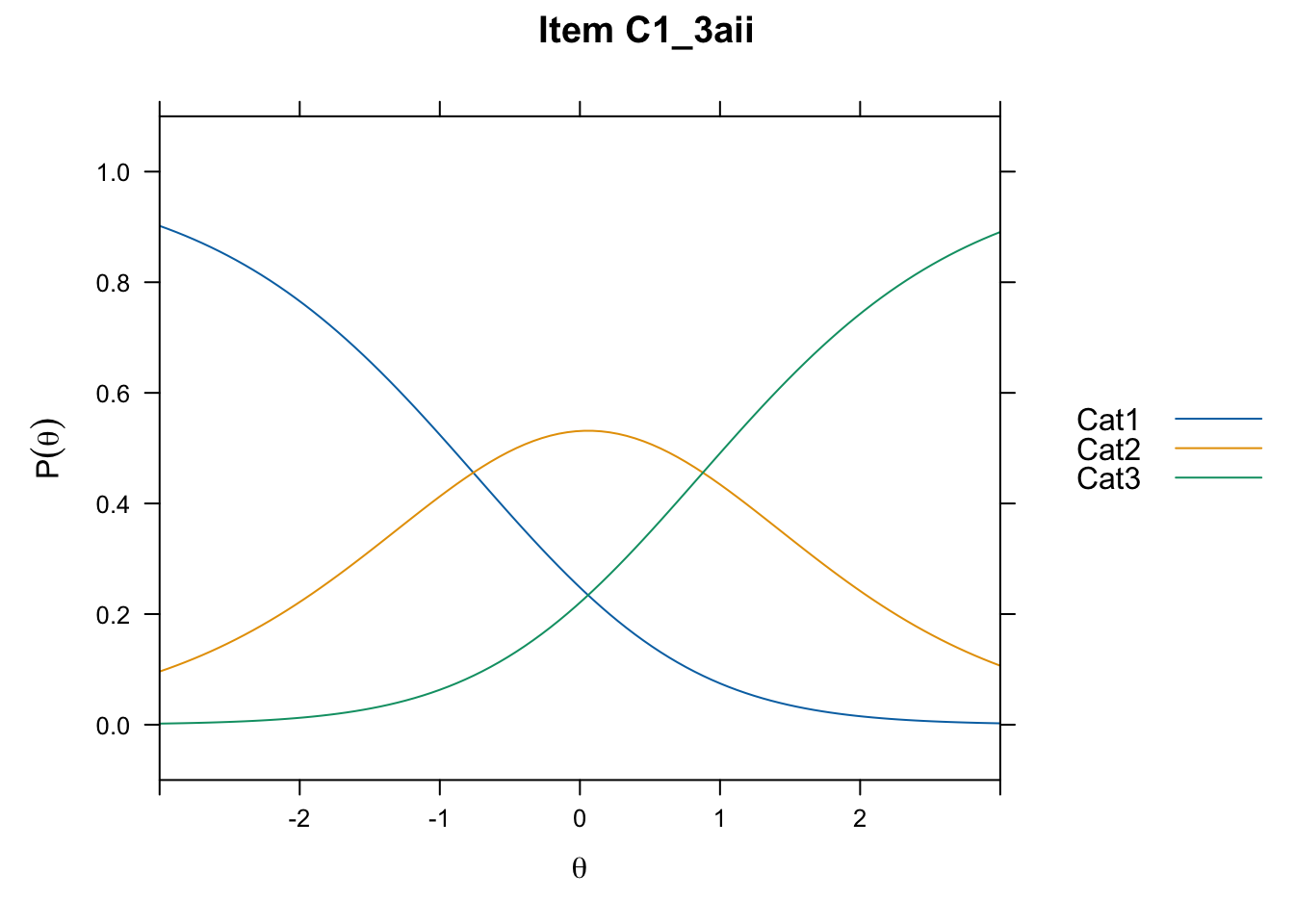
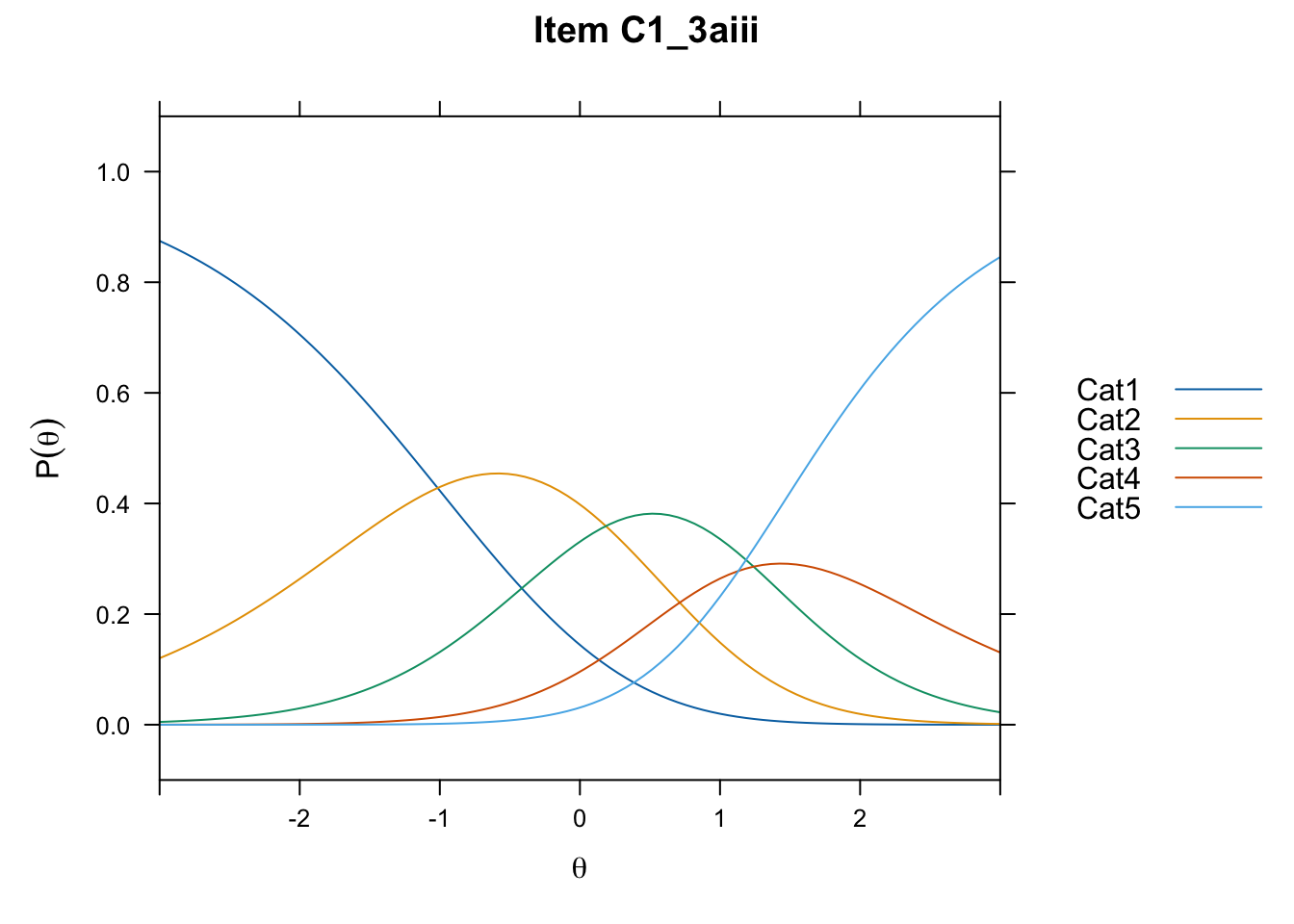
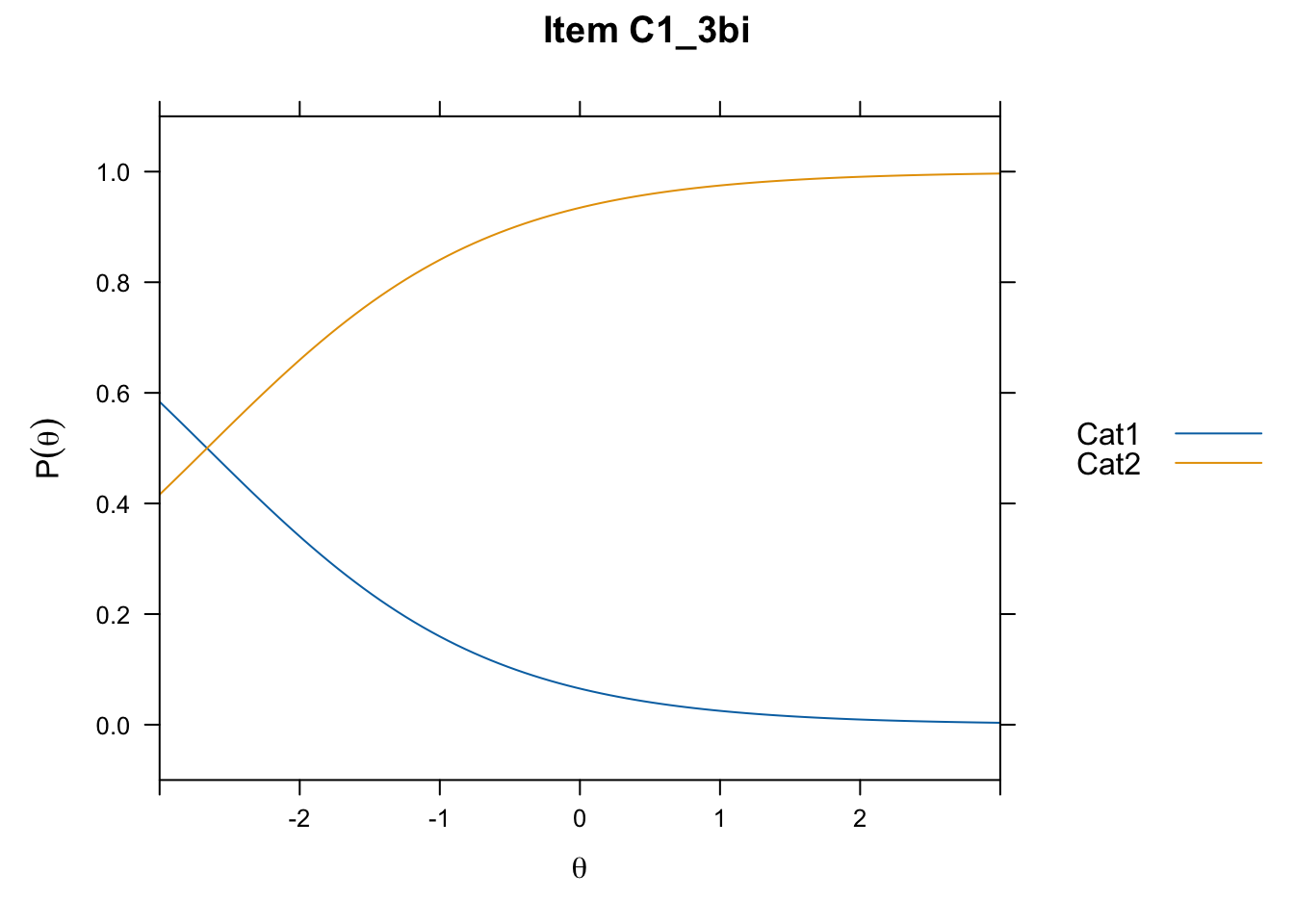
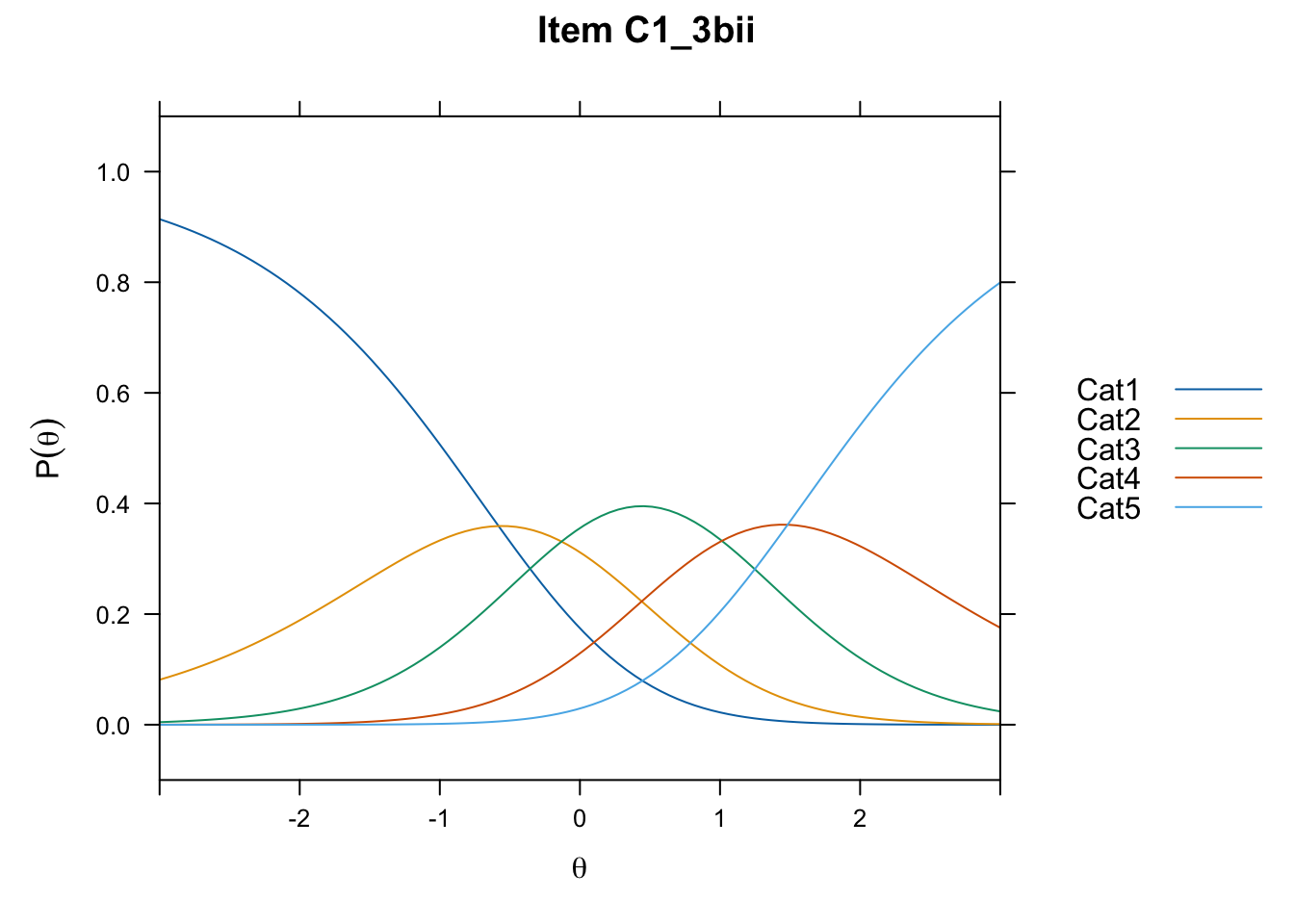
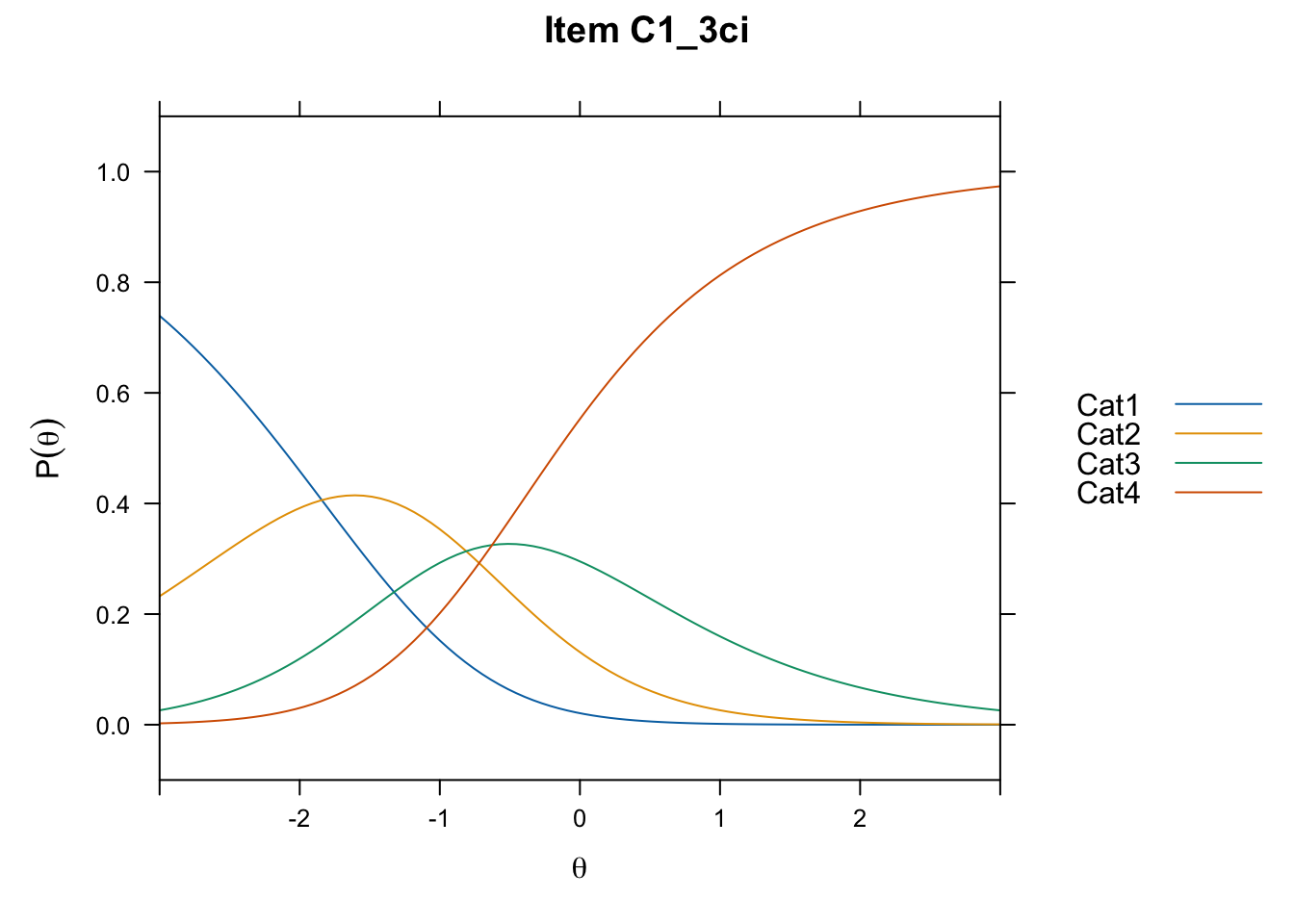
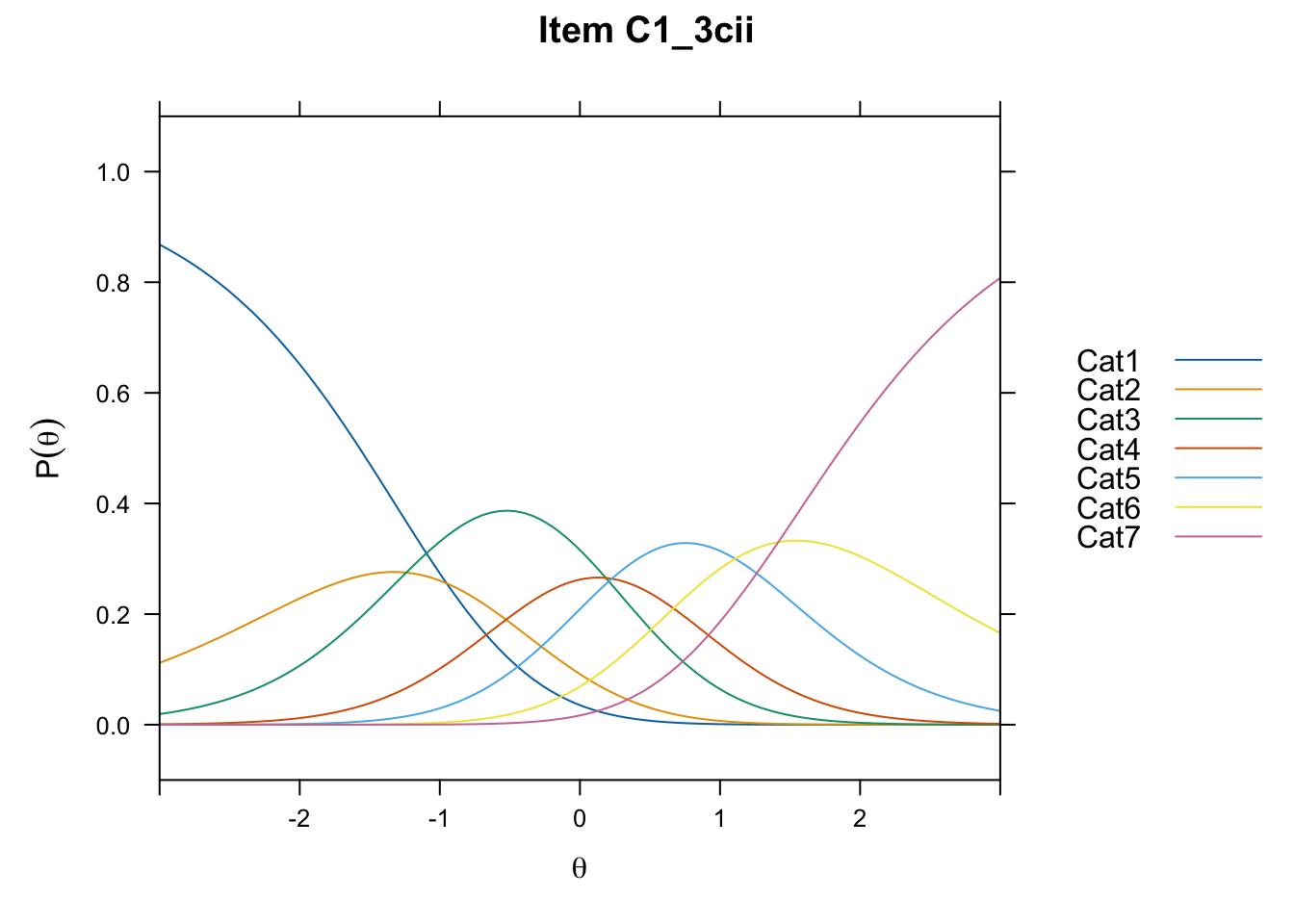
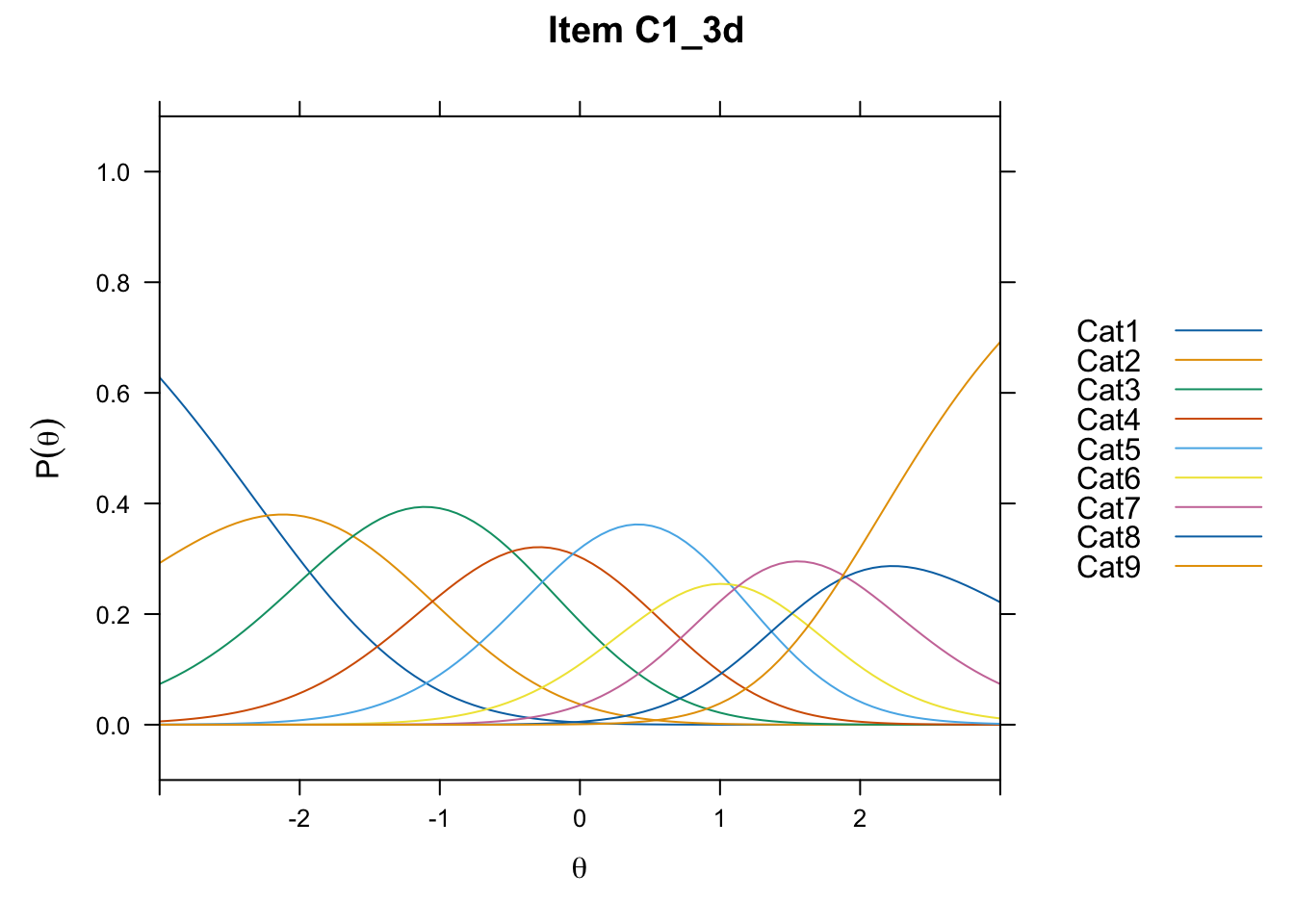
....................................................
Plots exported in png format into folder:
/Users/chris/Documents/CM3/Plotsknitr::kable(mod1$item1) #show item parameters (deltas)| xsi.label | xsi.index | xsi | se.xsi |
|---|---|---|---|
| C1_1ai_Cat1 | 1 | -3.4986161 | 0.0604511 |
| C1_1aii_Cat1 | 2 | -4.8625343 | 0.4173456 |
| C1_1aii_Cat2 | 3 | -2.8366876 | 0.0730070 |
| C1_1aii_Cat3 | 4 | -1.2655936 | 0.0267454 |
| C1_1bi_Cat1 | 5 | -3.6477143 | 0.0651326 |
| C1_1bii_Cat1 | 6 | -0.9767451 | 0.0254666 |
| C1_1biii_Cat1 | 7 | -0.4366899 | 0.0258152 |
| C1_1biii_Cat2 | 8 | 0.5419139 | 0.0259004 |
| C1_1biv_Cat1 | 9 | -0.2838066 | 0.0243368 |
| C1_1ci_Cat1 | 10 | 0.5102158 | 0.0266319 |
| C1_1ci_Cat2 | 11 | 0.4154921 | 0.0285255 |
| C1_1ci_Cat3 | 12 | 1.3819532 | 0.0392739 |
| C1_1ci_Cat4 | 13 | 1.6927569 | 0.0662257 |
| C1_1cii_Cat1 | 14 | -1.7685170 | 0.0531738 |
| C1_1cii_Cat2 | 15 | -1.0229517 | 0.0307266 |
| C1_1cii_Cat3 | 16 | 0.4905679 | 0.0257347 |
| C1_1cii_Cat4 | 17 | 0.5174543 | 0.0290481 |
| C1_1cii_Cat5 | 18 | 1.6122346 | 0.0437076 |
| C1_1cii_Cat6 | 19 | 1.9737824 | 0.0816198 |
| C1_1di_Cat1 | 20 | -0.3802452 | 0.0309360 |
| C1_1di_Cat2 | 21 | -0.5682350 | 0.0257794 |
| C1_1di_Cat3 | 22 | 0.6410594 | 0.0274623 |
| C1_1dii_Cat1 | 23 | -0.4634295 | 0.0275668 |
| C1_1dii_Cat2 | 24 | 0.0669450 | 0.0247250 |
| C1_1e_Cat1 | 25 | -2.4091577 | 0.0938733 |
| C1_1e_Cat2 | 26 | -1.5632376 | 0.0467979 |
| C1_1e_Cat3 | 27 | -0.6706033 | 0.0311514 |
| C1_1e_Cat4 | 28 | -0.1755785 | 0.0266092 |
| C1_1e_Cat5 | 29 | 0.6808900 | 0.0279851 |
| C1_1e_Cat6 | 30 | 0.9166584 | 0.0343270 |
| C1_1e_Cat7 | 31 | 1.5627424 | 0.0502252 |
| C1_1e_Cat8 | 32 | 1.8110832 | 0.0859609 |
| C1_1eSPaG_Cat1 | 33 | -1.3906254 | 0.0868852 |
| C1_1eSPaG_Cat2 | 34 | -2.7524203 | 0.0572619 |
| C1_1eSPaG_Cat3 | 35 | -0.8648779 | 0.0257222 |
| C1_1eSPaG_Cat4 | 36 | 2.0952676 | 0.0380979 |
| C1_2ai_Cat1 | 37 | -1.6071503 | 0.0588819 |
| C1_2ai_Cat2 | 38 | -1.7468042 | 0.0363285 |
| C1_2ai_Cat3 | 39 | 5.1313917 | 0.0264994 |
| C1_2ai_Cat4 | 40 | -5.3644469 | 0.0264885 |
| C1_2aii_Cat1 | 41 | -2.9870639 | 0.1357262 |
| C1_2aii_Cat2 | 42 | -2.1200165 | 0.0543546 |
| C1_2aii_Cat3 | 43 | -0.8253410 | 0.0287877 |
| C1_2aii_Cat4 | 44 | -0.0227188 | 0.0241951 |
| C1_2bi_Cat1 | 45 | -2.8938612 | 0.0512756 |
| C1_2bii_Cat1 | 46 | 0.0928013 | 0.0322967 |
| C1_2bii_Cat2 | 47 | -1.6797497 | 0.0272497 |
| C1_2biii_Cat1 | 48 | -1.7767989 | 0.1283571 |
| C1_2biii_Cat2 | 49 | -3.3948963 | 0.0750580 |
| C1_2biii_Cat3 | 50 | -0.8115457 | 0.0247937 |
| C1_2biv_Cat1 | 51 | 1.1174464 | 0.0274254 |
| C1_2biv_Cat2 | 52 | -1.6571716 | 0.0261064 |
| C1_2bv_Cat1 | 53 | 1.1732474 | 0.0254686 |
| C1_2bv_Cat2 | 54 | -0.0989439 | 0.0282216 |
| C1_2c_Cat1 | 55 | 0.5378311 | 0.0313037 |
| C1_2c_Cat2 | 56 | -0.2236799 | 0.0300944 |
| C1_2c_Cat3 | 57 | -0.3805059 | 0.0289878 |
| C1_2c_Cat4 | 58 | 0.3577677 | 0.0290282 |
| C1_2c_Cat5 | 59 | 0.9201111 | 0.0336968 |
| C1_2c_Cat6 | 60 | 1.2179333 | 0.0475174 |
| C1_2d_Cat1 | 61 | -2.2568439 | 0.0867304 |
| C1_2d_Cat2 | 62 | -1.4226014 | 0.0440320 |
| C1_2d_Cat3 | 63 | -0.5278258 | 0.0298808 |
| C1_2d_Cat4 | 64 | -0.1889719 | 0.0261161 |
| C1_2d_Cat5 | 65 | 1.2402257 | 0.0309157 |
| C1_2d_Cat6 | 66 | 1.1705637 | 0.0421757 |
| C1_2d_Cat7 | 67 | 2.0278678 | 0.0722968 |
| C1_2d_Cat8 | 68 | 2.0862339 | 0.1343542 |
| C1_3ai_Cat1 | 69 | -2.5341704 | 0.0758054 |
| C1_3ai_Cat2 | 70 | -0.8667725 | 0.0354036 |
| C1_3ai_Cat3 | 71 | -0.4616945 | 0.0272621 |
| C1_3ai_Cat4 | 72 | -0.2272080 | 0.0248870 |
| C1_3aii_Cat1 | 73 | -0.7609214 | 0.0269400 |
| C1_3aii_Cat2 | 74 | 0.8778269 | 0.0269865 |
| C1_3aiii_Cat1 | 75 | -1.0155021 | 0.0346077 |
| C1_3aiii_Cat2 | 76 | 0.1851119 | 0.0263538 |
| C1_3aiii_Cat3 | 77 | 1.2395251 | 0.0323114 |
| C1_3aiii_Cat4 | 78 | 1.1316478 | 0.0465848 |
| C1_3bi_Cat1 | 79 | -2.6614924 | 0.0429139 |
| C1_3bii_Cat1 | 80 | -0.5806264 | 0.0301459 |
| C1_3bii_Cat2 | 81 | -0.1311194 | 0.0253035 |
| C1_3bii_Cat3 | 82 | 1.0133880 | 0.0296646 |
| C1_3bii_Cat4 | 83 | 1.4827345 | 0.0473044 |
| C1_3ci_Cat1 | 84 | -1.8421460 | 0.0569711 |
| C1_3ci_Cat2 | 85 | -0.8114687 | 0.0316059 |
| C1_3ci_Cat3 | 86 | -0.6268538 | 0.0245684 |
| C1_3cii_Cat1 | 87 | -0.9514191 | 0.0494433 |
| C1_3cii_Cat2 | 88 | -1.2393889 | 0.0353659 |
| C1_3cii_Cat3 | 89 | 0.1849122 | 0.0274597 |
| C1_3cii_Cat4 | 90 | 0.2278265 | 0.0277558 |
| C1_3cii_Cat5 | 91 | 1.1078550 | 0.0343116 |
| C1_3cii_Cat6 | 92 | 1.4157452 | 0.0524217 |
| C1_3d_Cat1 | 93 | -2.2354330 | 0.0907095 |
| C1_3d_Cat2 | 94 | -1.6195972 | 0.0461023 |
| C1_3d_Cat3 | 95 | -0.4846092 | 0.0296708 |
| C1_3d_Cat4 | 96 | -0.0501634 | 0.0262860 |
| C1_3d_Cat5 | 97 | 1.0723274 | 0.0304509 |
| C1_3d_Cat6 | 98 | 1.1322721 | 0.0404484 |
| C1_3d_Cat7 | 99 | 1.8968665 | 0.0651859 |
| C1_3d_Cat8 | 100 | 1.8606380 | 0.1121267 |
28.2 Things to consider
Are there items with disordered thresholds? Why might this be? Are there any misfitting items? Why might this be? How well targeted is the test?
28.3 Extension exercises
Try CM2 and CM3 by changing the code to select items that start with C2_ or C3_
resp_c2 <- resp %>%
select(starts_with('C2_'))