rm(list=ls())
library(tidyverse)
library(dexter)
# load in the dataset
responses <- read_csv('data/pc-data.csv')
# recode NA as 0 across all columns
responses <- responses %>%
mutate_if(is.numeric, ~replace_na(., 0))
# load the data summary
data_summary <- read_csv('data/pc-data-summary.csv')27 Fitting the Partial Credit Model
You can fit the Partial Credit Model in either dexter or TAM. Let’s start with dexter.
# Create the rules from the summary
# create an empty tibble with three columns item_id, response, and item_score
rules <- tibble(item_id = character(), response = character(), item_score = numeric())
# for each row in the data summary create a rule
for (i in 1:nrow(data_summary)) {
# get the item id
item_id <- data_summary$Variable[i]
# get the maximum mark
max_mark <- data_summary$`Maximum possible mark`[i]
# for every mark from 0 to max_mark create a rule
for (mark in 0:max_mark) {
# create a rule for the mark
rule <- tibble(item_id = item_id, response = as.character(mark), item_score = mark)
# add the rule to the rules tibble
rules <- bind_rows(rules, rule)
}
}# create a new project
db <- start_new_project(rules, db_name = ":memory:", person_properties=list(sex="unknown",grade="unknown"))
# add the response data
add_booklet(db, responses, "geography")no column `person_id` provided, automatically generating unique person id's$items
[1] "C1_1ai" "C1_1aii" "C1_1bi" "C1_1bii" "C1_1biii" "C1_1biv"
[7] "C1_1ci" "C1_1cii" "C1_1di" "C1_1dii" "C1_1e" "C1_1eSPaG"
[13] "C1_2ai" "C1_2aii" "C1_2bi" "C1_2bii" "C1_2biii" "C1_2biv"
[19] "C1_2bv" "C1_2c" "C1_2d" "C1_3ai" "C1_3aii" "C1_3aiii"
[25] "C1_3bi" "C1_3bii" "C1_3ci" "C1_3cii" "C1_3d" "C2_Aa"
[31] "C2_Ab" "C2_Ac" "C2_Adi" "C2_Adii" "C2_Aei" "C2_Aeii"
[37] "C2_Aeiii" "C2_Aeiv" "C2_Af" "C2_Ag" "C2_Bai" "C2_Baii"
[43] "C2_Bb" "C2_Bc" "C2_Bd" "C2_Be" "C2_Bf" "C2_C"
[49] "C2_CSPaG" "C3_1ai" "C3_1aii" "C3_1bi" "C3_1bii" "C3_1biii"
[55] "C3_1c" "C3_2ai" "C3_2aii" "C3_2b" "C3_2ci" "C3_2cii"
[61] "C3_2ciii" "C3_2d" "C3_3ai" "C3_3aii" "C3_3aiii" "C3_3b"
[67] "C3_3ci" "C3_3cii" "C3_3di" "C3_3dii" "C3_3e" "C3_3f"
[73] "C3_3fSPaG"
$person_properties
[1] "sex" "grade"
$columns_ignored
[1] "C1_total" "C2_total" "C3_total" "total" # print the item tables
tt = tia_tables(db)
knitr::kable(tt$items, digits=2)| booklet_id | item_id | mean_score | sd_score | max_score | pvalue | rit | rir | n_persons |
|---|---|---|---|---|---|---|---|---|
| geography | C1_1ai | 0.96 | 0.18 | 1 | 0.96 | 0.24 | 0.24 | 8500 |
| geography | C1_1aii | 2.72 | 0.51 | 3 | 0.91 | 0.38 | 0.37 | 8500 |
| geography | C1_1bi | 0.97 | 0.18 | 1 | 0.97 | 0.20 | 0.19 | 8500 |
| geography | C1_1bii | 0.71 | 0.45 | 1 | 0.71 | 0.34 | 0.33 | 8500 |
| geography | C1_1biii | 0.98 | 0.78 | 2 | 0.49 | 0.43 | 0.41 | 8500 |
| geography | C1_1biv | 0.53 | 0.50 | 1 | 0.53 | 0.49 | 0.48 | 8500 |
| geography | C1_1ci | 0.91 | 1.12 | 4 | 0.23 | 0.46 | 0.44 | 8500 |
| geography | C1_1cii | 2.50 | 1.40 | 6 | 0.42 | 0.61 | 0.58 | 8500 |
| geography | C1_1di | 1.63 | 1.08 | 3 | 0.54 | 0.44 | 0.42 | 8500 |
| geography | C1_1dii | 1.09 | 0.81 | 2 | 0.54 | 0.49 | 0.47 | 8500 |
| geography | C1_1e | 3.75 | 1.73 | 8 | 0.47 | 0.69 | 0.67 | 8500 |
| geography | C1_1eSPaG | 2.69 | 0.80 | 4 | 0.67 | 0.60 | 0.59 | 8500 |
| geography | C1_2ai | 2.85 | 1.28 | 4 | 0.71 | 0.49 | 0.46 | 8500 |
| geography | C1_2aii | 3.12 | 0.92 | 4 | 0.78 | 0.35 | 0.32 | 8500 |
| geography | C1_2bi | 0.85 | 0.36 | 1 | 0.85 | 0.36 | 0.35 | 8500 |
| geography | C1_2bii | 1.43 | 0.84 | 2 | 0.71 | 0.46 | 0.44 | 8500 |
| geography | C1_2biii | 2.61 | 0.62 | 3 | 0.87 | 0.38 | 0.37 | 8500 |
| geography | C1_2biv | 1.15 | 0.95 | 2 | 0.58 | 0.57 | 0.55 | 8500 |
| geography | C1_2bv | 0.65 | 0.85 | 2 | 0.33 | 0.50 | 0.49 | 8500 |
| geography | C1_2c | 2.36 | 2.01 | 6 | 0.39 | 0.68 | 0.65 | 8500 |
| geography | C1_2d | 3.34 | 1.65 | 8 | 0.42 | 0.67 | 0.65 | 8500 |
| geography | C1_3ai | 2.90 | 1.16 | 4 | 0.72 | 0.65 | 0.64 | 8500 |
| geography | C1_3aii | 0.96 | 0.73 | 2 | 0.48 | 0.37 | 0.36 | 8500 |
| geography | C1_3aiii | 1.41 | 1.19 | 4 | 0.35 | 0.54 | 0.51 | 8500 |
| geography | C1_3bi | 0.92 | 0.28 | 1 | 0.92 | 0.28 | 0.28 | 8500 |
| geography | C1_3bii | 1.57 | 1.18 | 4 | 0.39 | 0.54 | 0.51 | 8500 |
| geography | C1_3ci | 2.24 | 0.96 | 3 | 0.75 | 0.60 | 0.58 | 8500 |
| geography | C1_3cii | 2.68 | 1.72 | 6 | 0.45 | 0.70 | 0.67 | 8500 |
| geography | C1_3d | 3.36 | 1.70 | 8 | 0.42 | 0.68 | 0.66 | 8500 |
| geography | C2_Aa | 2.34 | 1.27 | 5 | 0.47 | 0.40 | 0.37 | 8500 |
| geography | C2_Ab | 1.75 | 1.18 | 4 | 0.44 | 0.41 | 0.39 | 8500 |
| geography | C2_Ac | 2.83 | 1.15 | 4 | 0.71 | 0.56 | 0.54 | 8500 |
| geography | C2_Adi | 3.31 | 1.03 | 4 | 0.83 | 0.50 | 0.48 | 8500 |
| geography | C2_Adii | 1.42 | 0.93 | 3 | 0.47 | 0.51 | 0.50 | 8500 |
| geography | C2_Aei | 0.94 | 0.24 | 1 | 0.94 | 0.21 | 0.21 | 8500 |
| geography | C2_Aeii | 1.92 | 1.05 | 3 | 0.64 | 0.64 | 0.63 | 8500 |
| geography | C2_Aeiii | 0.78 | 0.85 | 4 | 0.20 | 0.41 | 0.40 | 8500 |
| geography | C2_Aeiv | 0.55 | 0.67 | 2 | 0.28 | 0.26 | 0.25 | 8500 |
| geography | C2_Af | 1.48 | 0.81 | 3 | 0.49 | 0.46 | 0.44 | 8500 |
| geography | C2_Ag | 3.17 | 1.52 | 6 | 0.53 | 0.66 | 0.64 | 8500 |
| geography | C2_Bai | 0.90 | 0.30 | 1 | 0.90 | 0.33 | 0.32 | 8500 |
| geography | C2_Baii | 0.77 | 0.57 | 2 | 0.38 | 0.49 | 0.48 | 8500 |
| geography | C2_Bb | 1.25 | 1.23 | 4 | 0.31 | 0.43 | 0.41 | 8500 |
| geography | C2_Bc | 1.11 | 0.75 | 2 | 0.56 | 0.40 | 0.39 | 8500 |
| geography | C2_Bd | 2.67 | 1.39 | 5 | 0.53 | 0.65 | 0.63 | 8500 |
| geography | C2_Be | 2.30 | 1.43 | 4 | 0.58 | 0.59 | 0.56 | 8500 |
| geography | C2_Bf | 1.21 | 0.97 | 3 | 0.40 | 0.50 | 0.49 | 8500 |
| geography | C2_C | 6.69 | 3.23 | 12 | 0.56 | 0.77 | 0.73 | 8500 |
| geography | C2_CSPaG | 2.95 | 1.06 | 4 | 0.74 | 0.70 | 0.69 | 8500 |
| geography | C3_1ai | 0.42 | 0.63 | 2 | 0.21 | 0.46 | 0.45 | 8500 |
| geography | C3_1aii | 1.70 | 0.61 | 2 | 0.85 | 0.44 | 0.42 | 8500 |
| geography | C3_1bi | 1.60 | 0.77 | 2 | 0.80 | 0.51 | 0.50 | 8500 |
| geography | C3_1bii | 0.72 | 0.74 | 2 | 0.36 | 0.46 | 0.44 | 8500 |
| geography | C3_1biii | 1.34 | 1.16 | 4 | 0.33 | 0.60 | 0.58 | 8500 |
| geography | C3_1c | 1.54 | 1.57 | 6 | 0.26 | 0.60 | 0.57 | 8500 |
| geography | C3_2ai | 1.64 | 0.59 | 2 | 0.82 | 0.16 | 0.15 | 8500 |
| geography | C3_2aii | 1.27 | 0.98 | 4 | 0.32 | 0.49 | 0.48 | 8500 |
| geography | C3_2b | 1.56 | 0.59 | 2 | 0.78 | 0.41 | 0.40 | 8500 |
| geography | C3_2ci | 0.77 | 0.42 | 1 | 0.77 | 0.37 | 0.36 | 8500 |
| geography | C3_2cii | 0.95 | 0.22 | 1 | 0.95 | 0.27 | 0.26 | 8500 |
| geography | C3_2ciii | 1.47 | 0.96 | 4 | 0.37 | 0.54 | 0.52 | 8500 |
| geography | C3_2d | 1.37 | 1.15 | 4 | 0.34 | 0.61 | 0.59 | 8500 |
| geography | C3_3ai | 2.43 | 0.75 | 3 | 0.81 | 0.50 | 0.48 | 8500 |
| geography | C3_3aii | 1.90 | 0.37 | 2 | 0.95 | 0.39 | 0.38 | 8500 |
| geography | C3_3aiii | 2.78 | 1.19 | 4 | 0.69 | 0.67 | 0.65 | 8500 |
| geography | C3_3b | 0.79 | 0.41 | 1 | 0.79 | 0.37 | 0.36 | 8500 |
| geography | C3_3ci | 1.39 | 0.87 | 2 | 0.70 | 0.42 | 0.40 | 8500 |
| geography | C3_3cii | 1.81 | 0.50 | 2 | 0.91 | 0.42 | 0.41 | 8500 |
| geography | C3_3di | 0.26 | 0.44 | 1 | 0.26 | 0.23 | 0.22 | 8500 |
| geography | C3_3dii | 2.29 | 1.58 | 6 | 0.38 | 0.62 | 0.59 | 8500 |
| geography | C3_3e | 0.95 | 0.81 | 3 | 0.32 | 0.44 | 0.43 | 8500 |
| geography | C3_3f | 5.55 | 2.64 | 12 | 0.46 | 0.75 | 0.72 | 8500 |
| geography | C3_3fSPaG | 2.88 | 1.05 | 4 | 0.72 | 0.64 | 0.62 | 8500 |
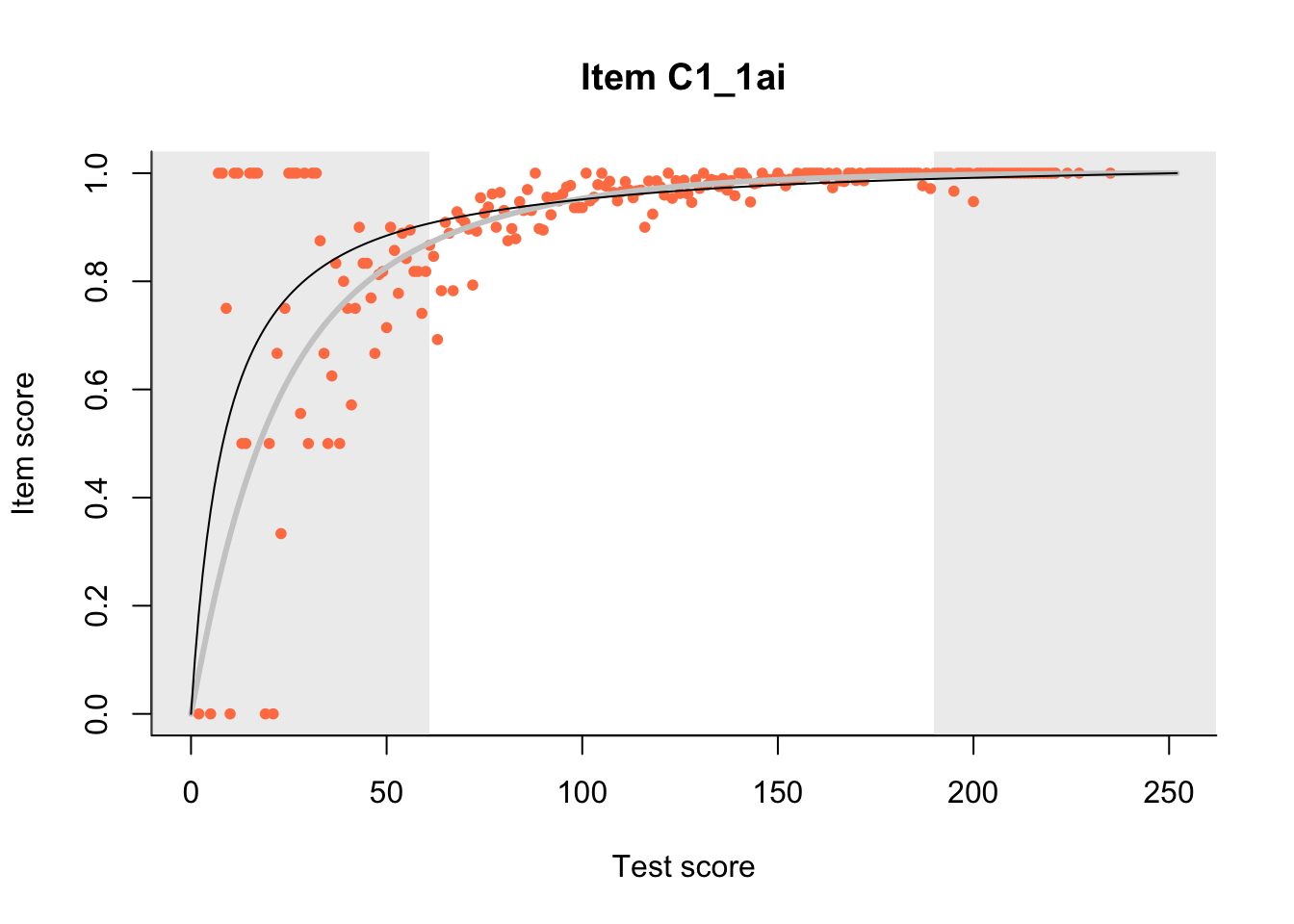
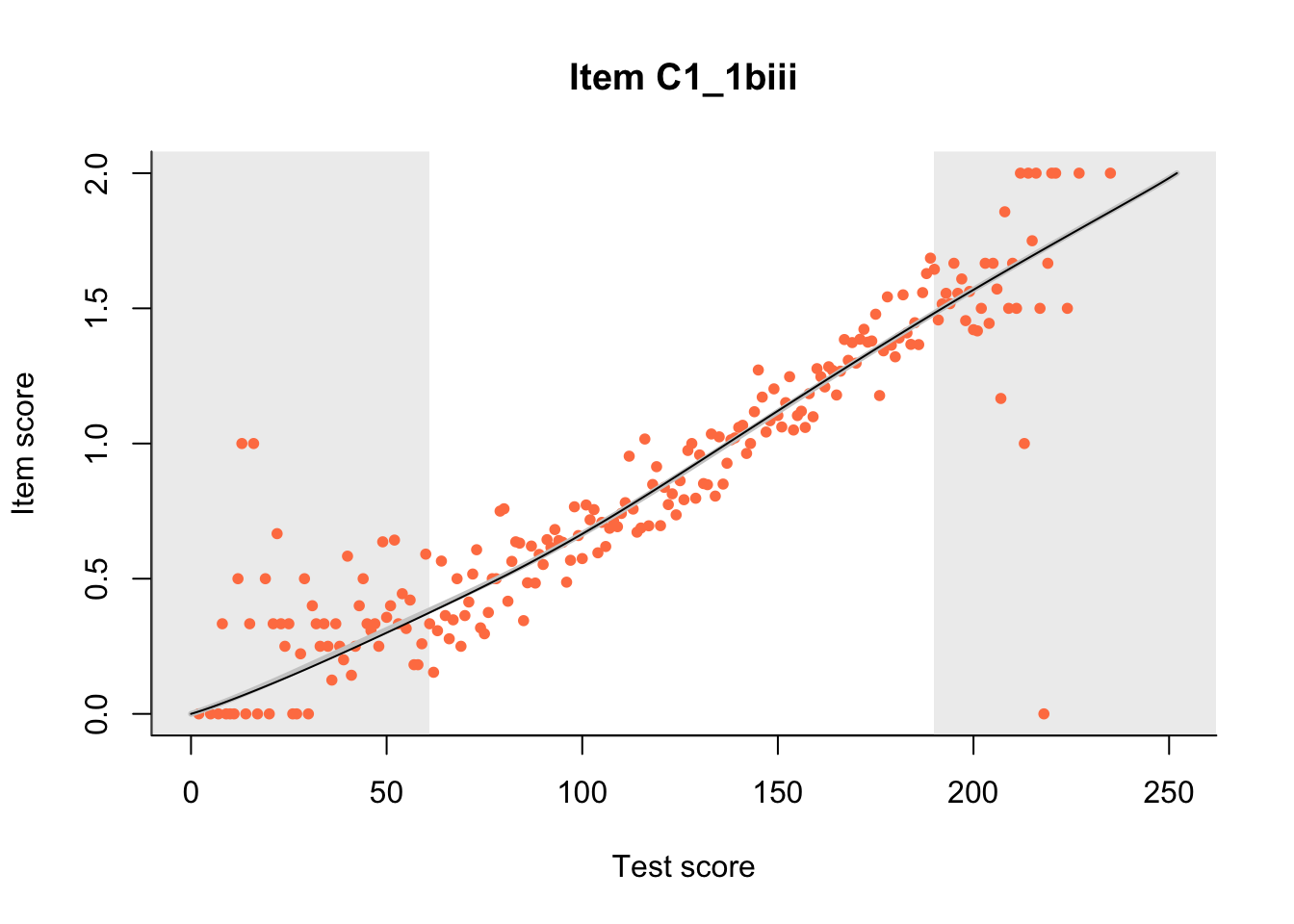
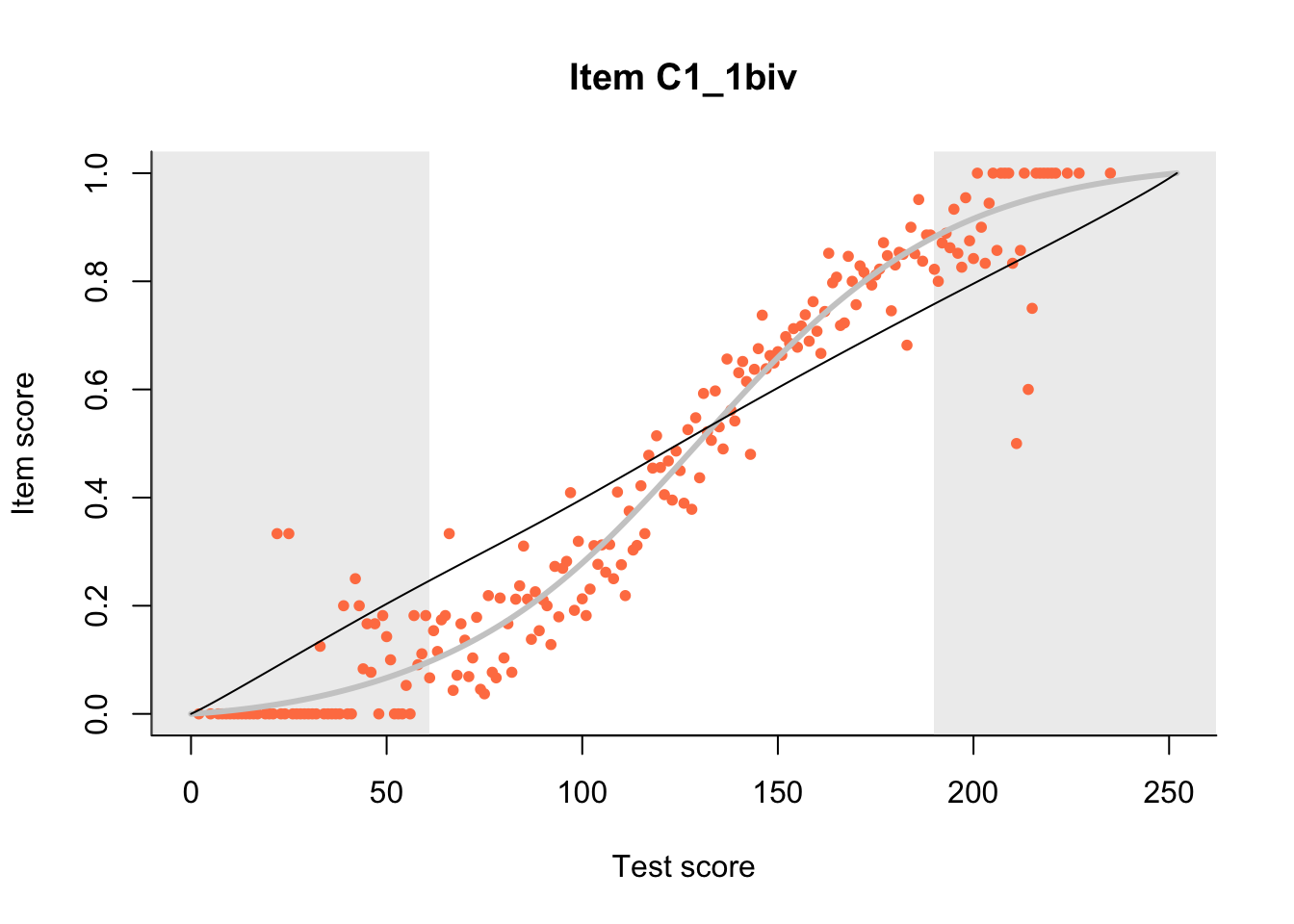
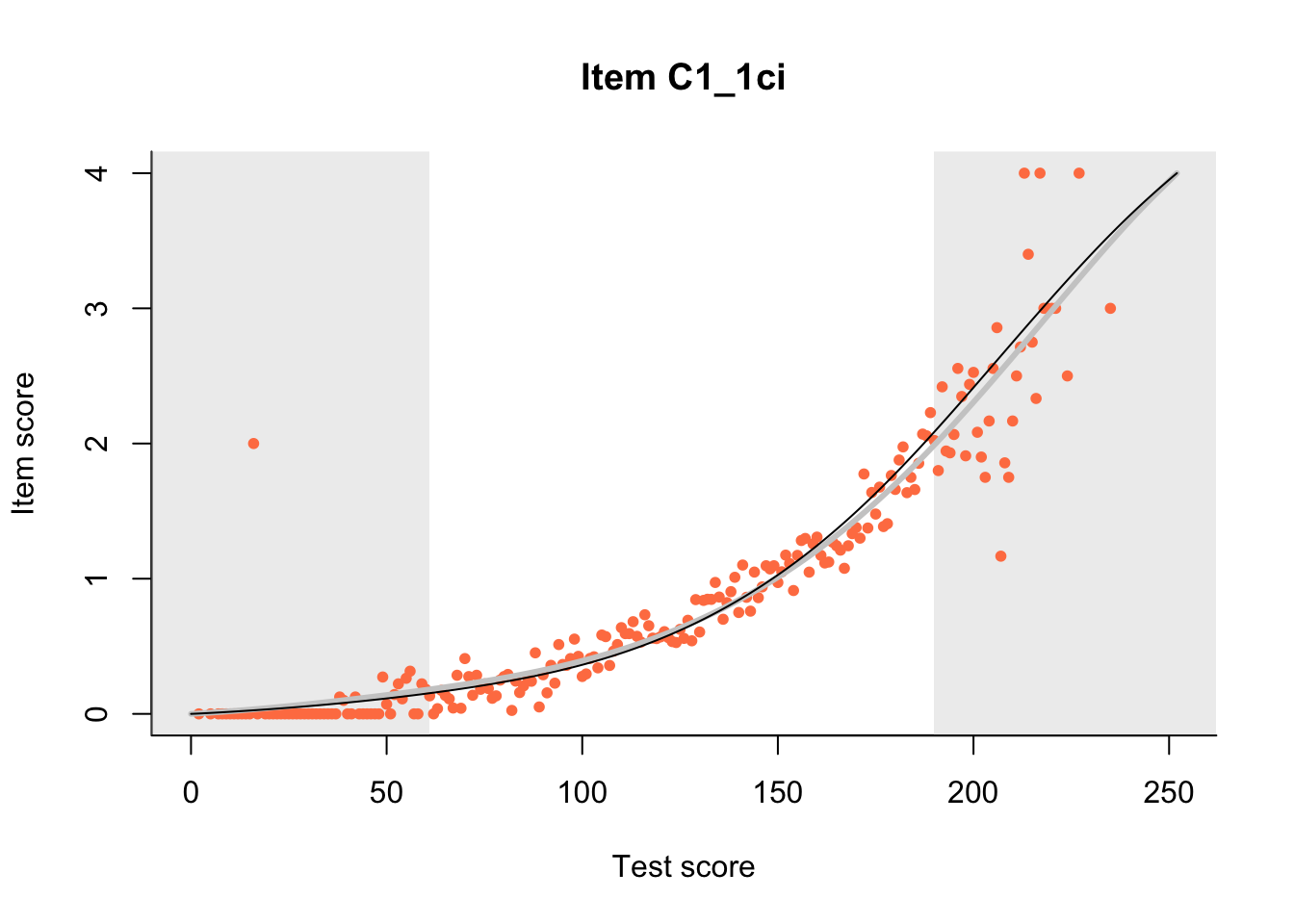
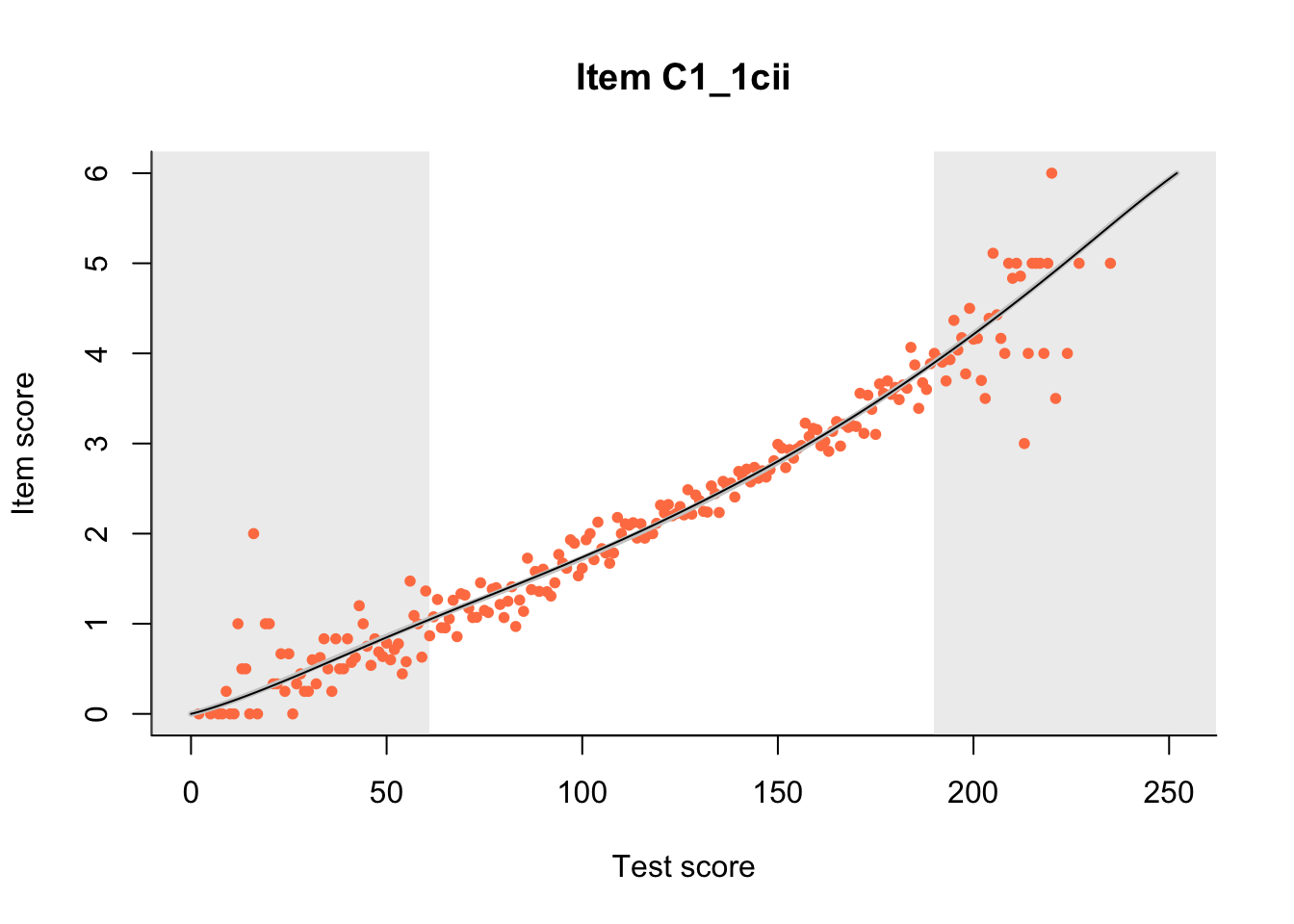
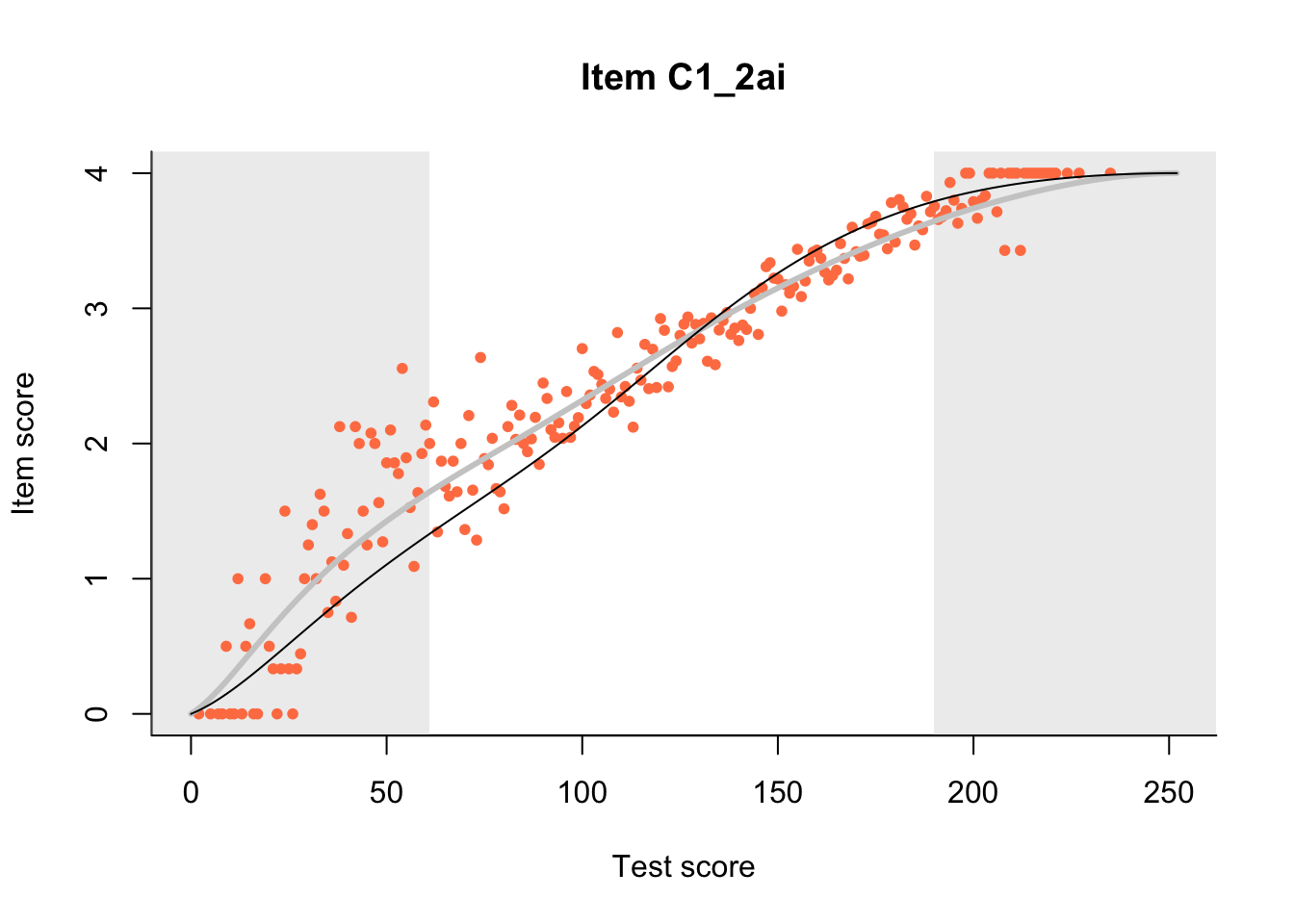
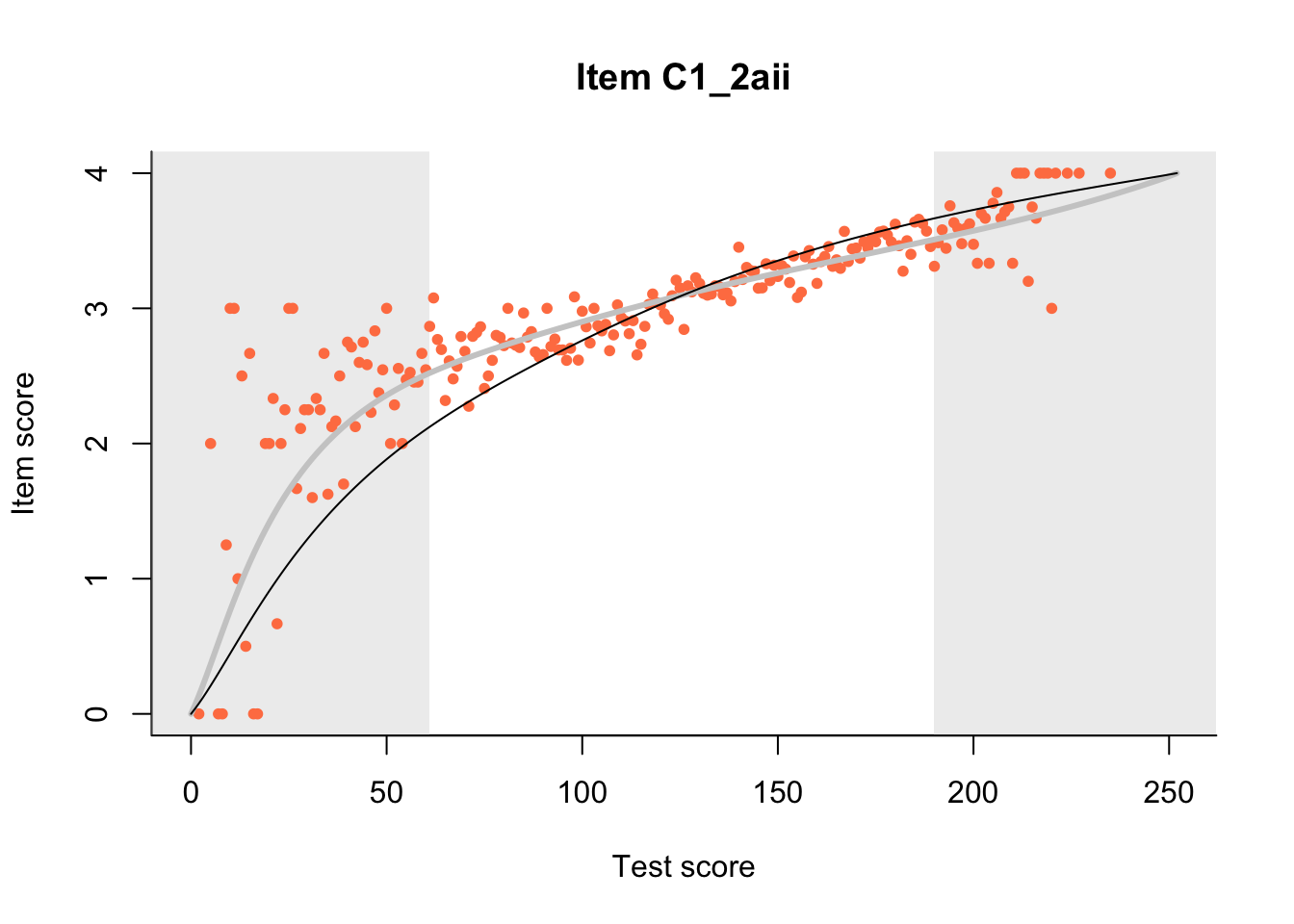
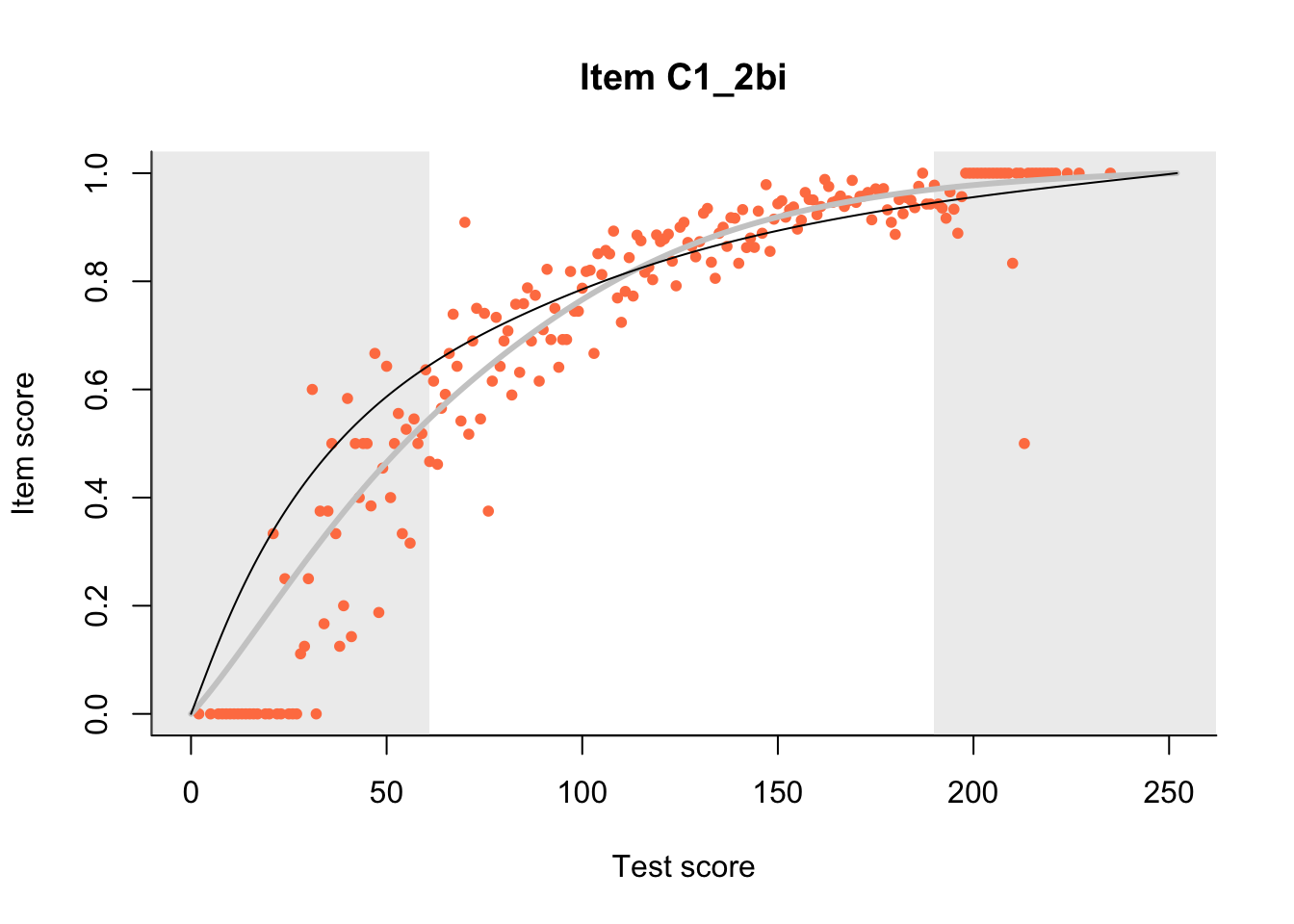
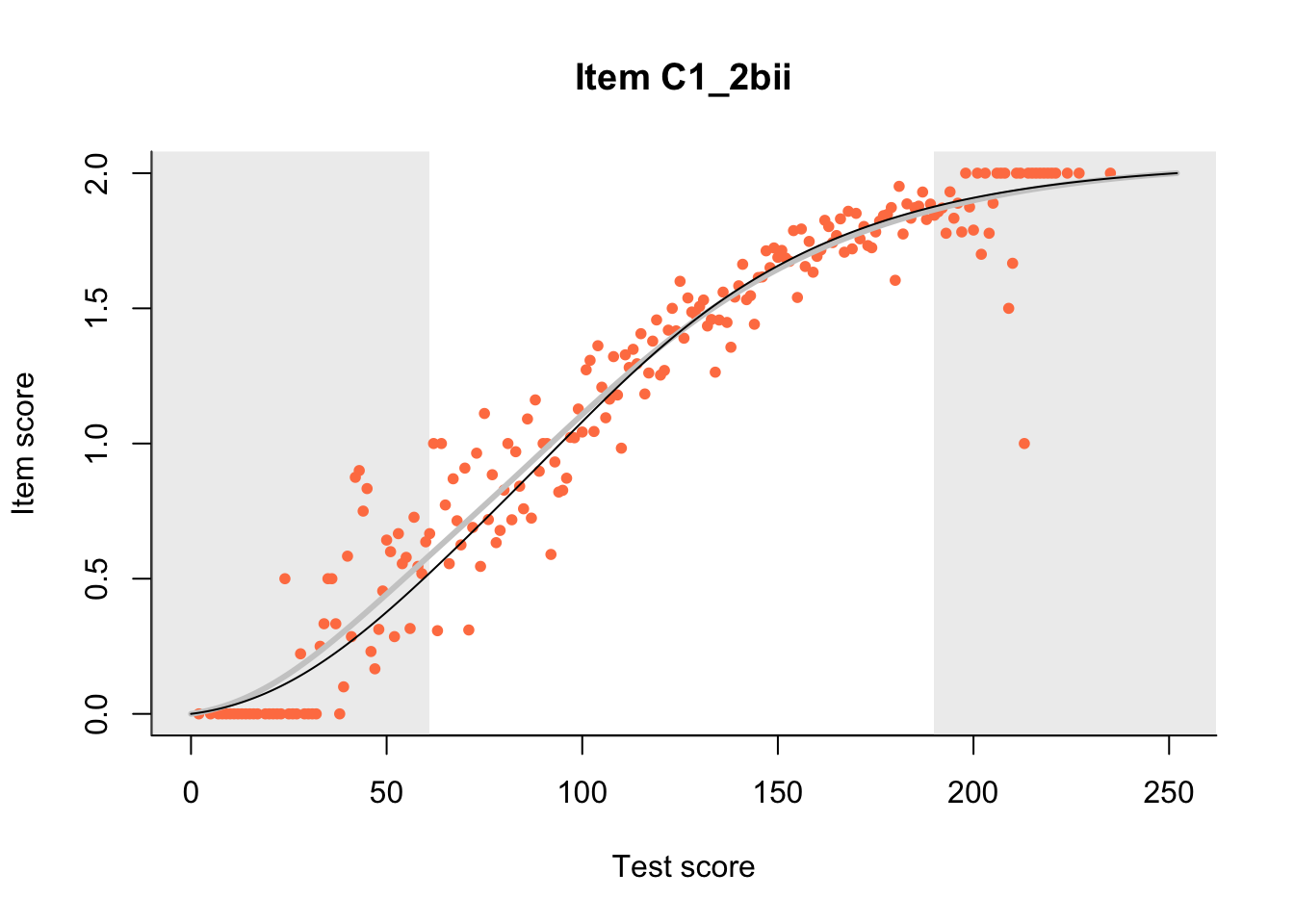
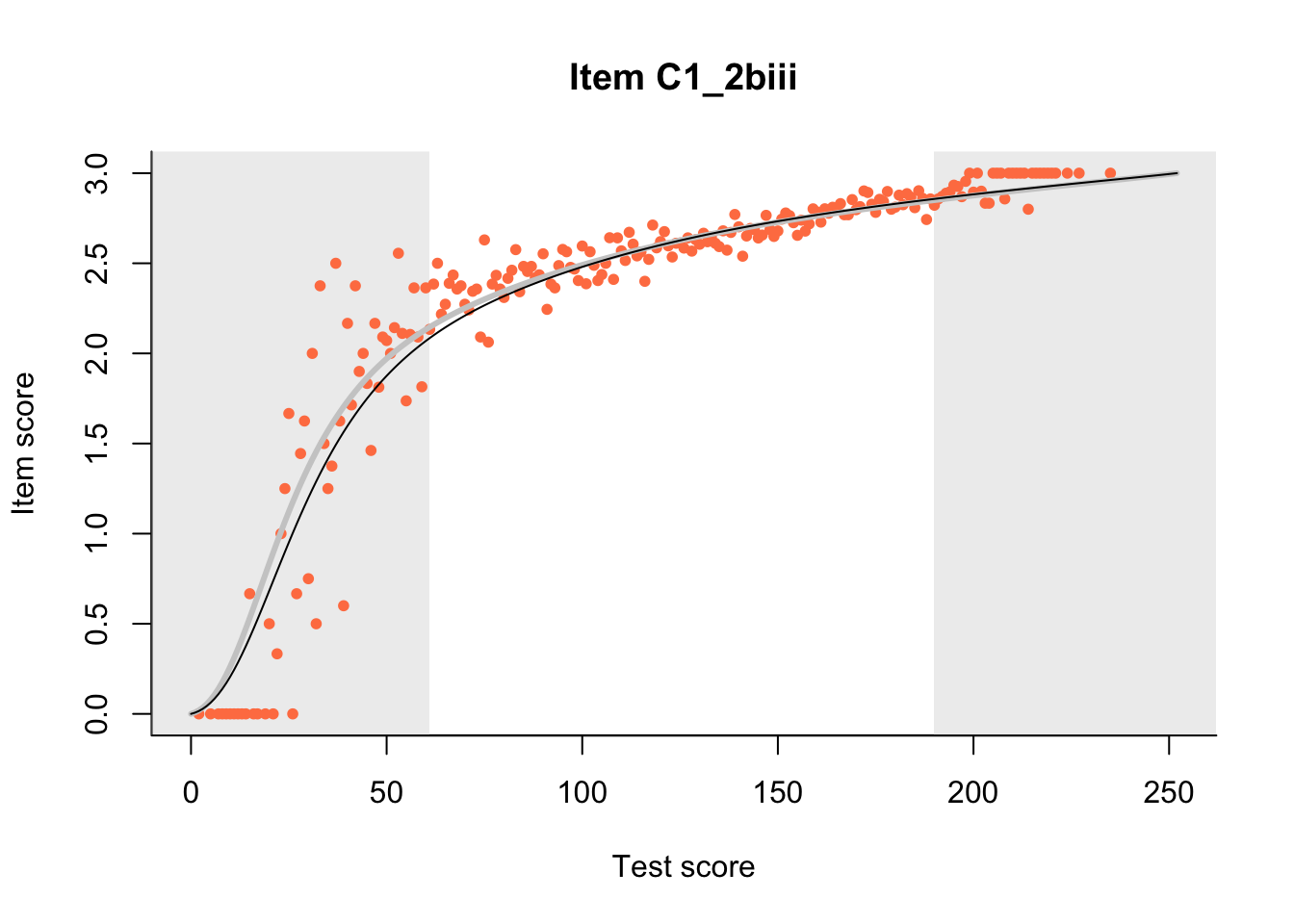
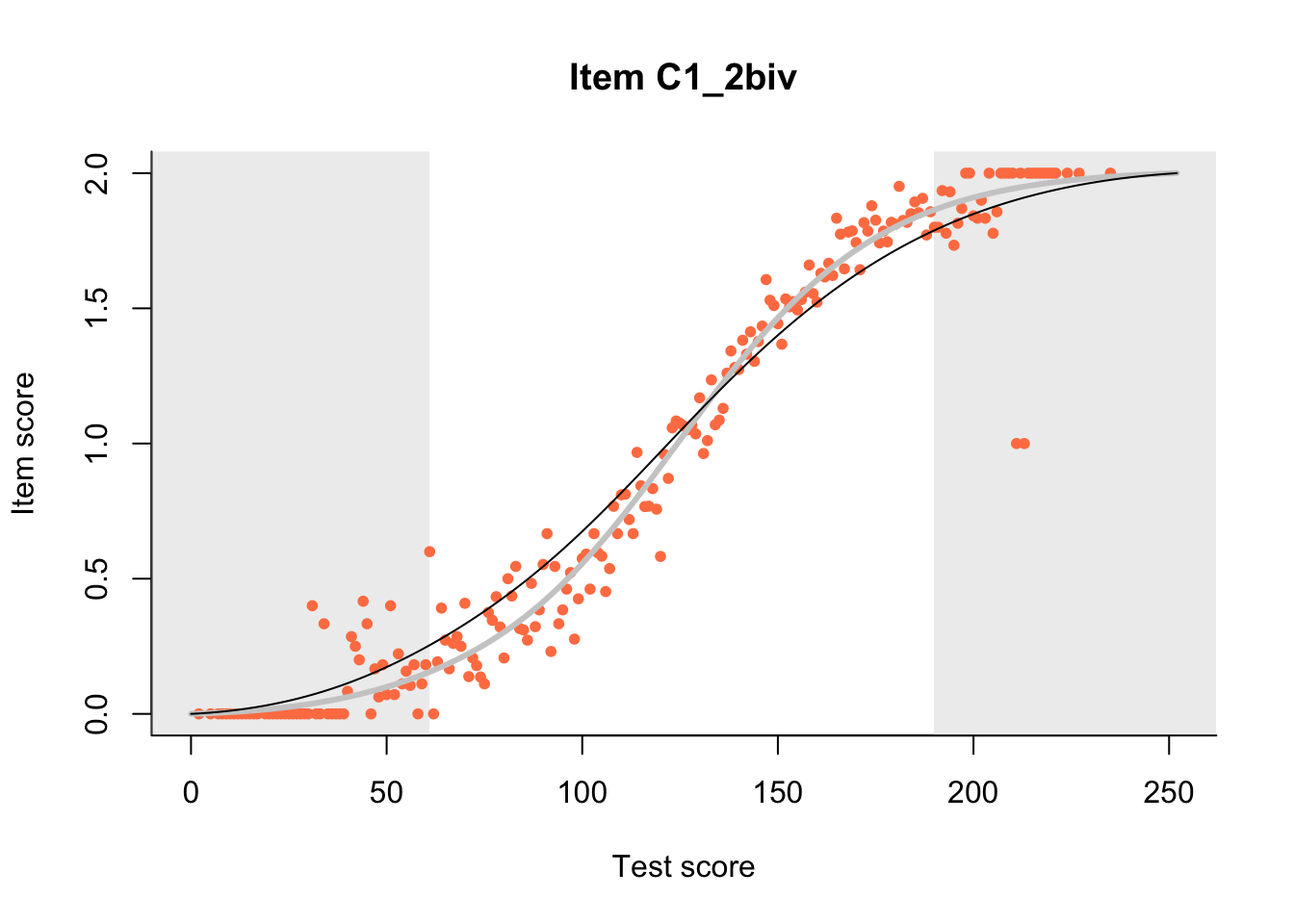
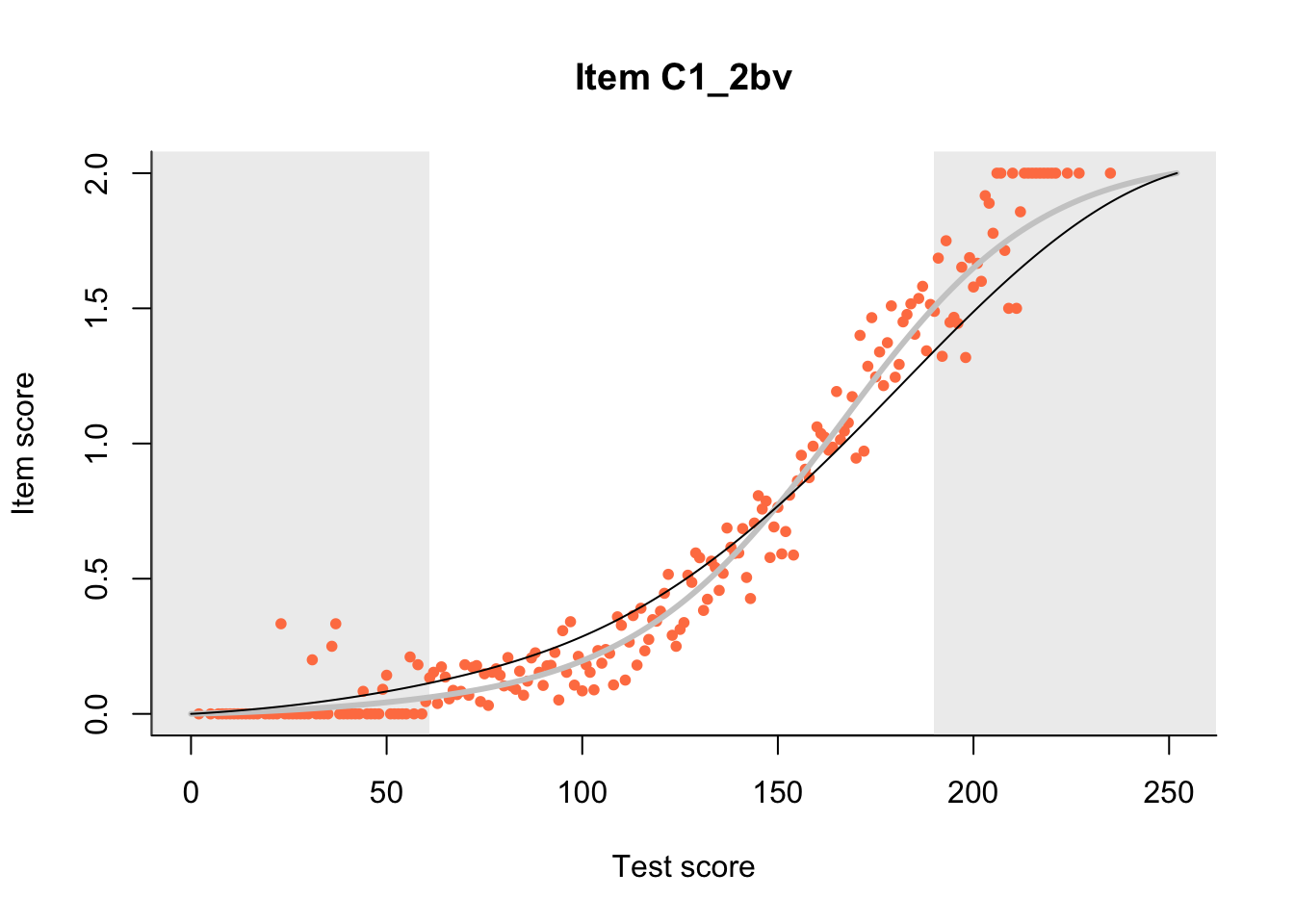
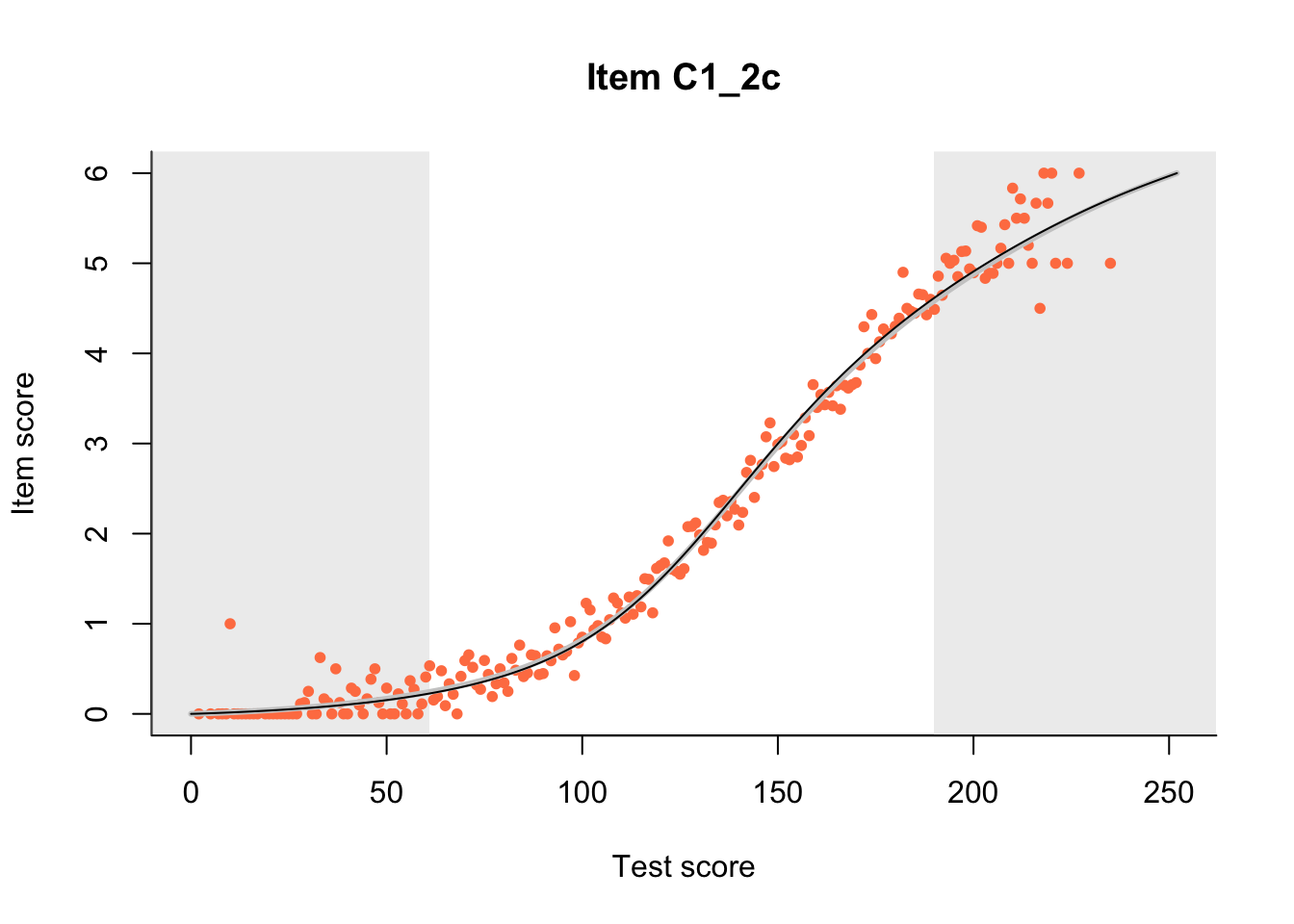
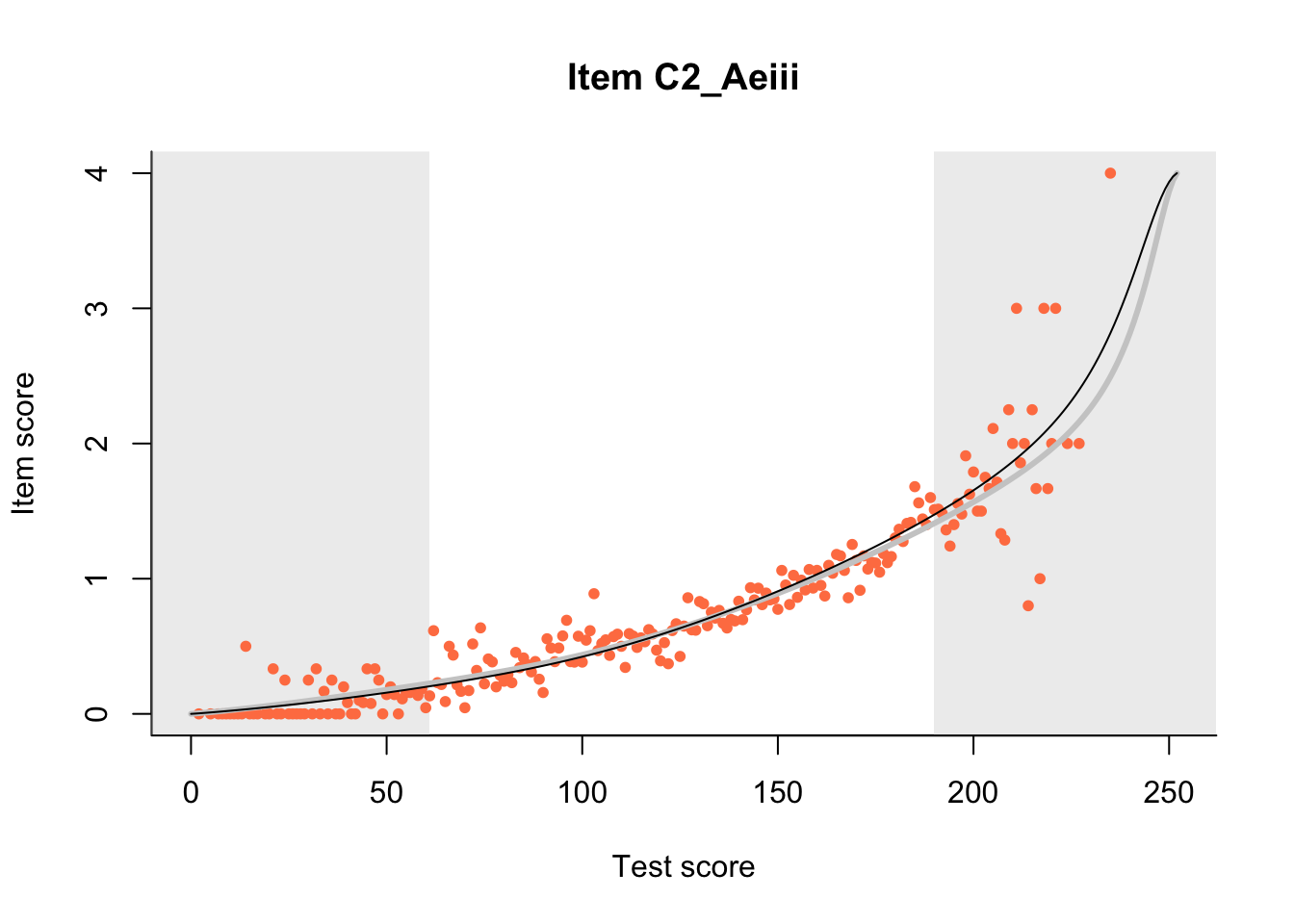
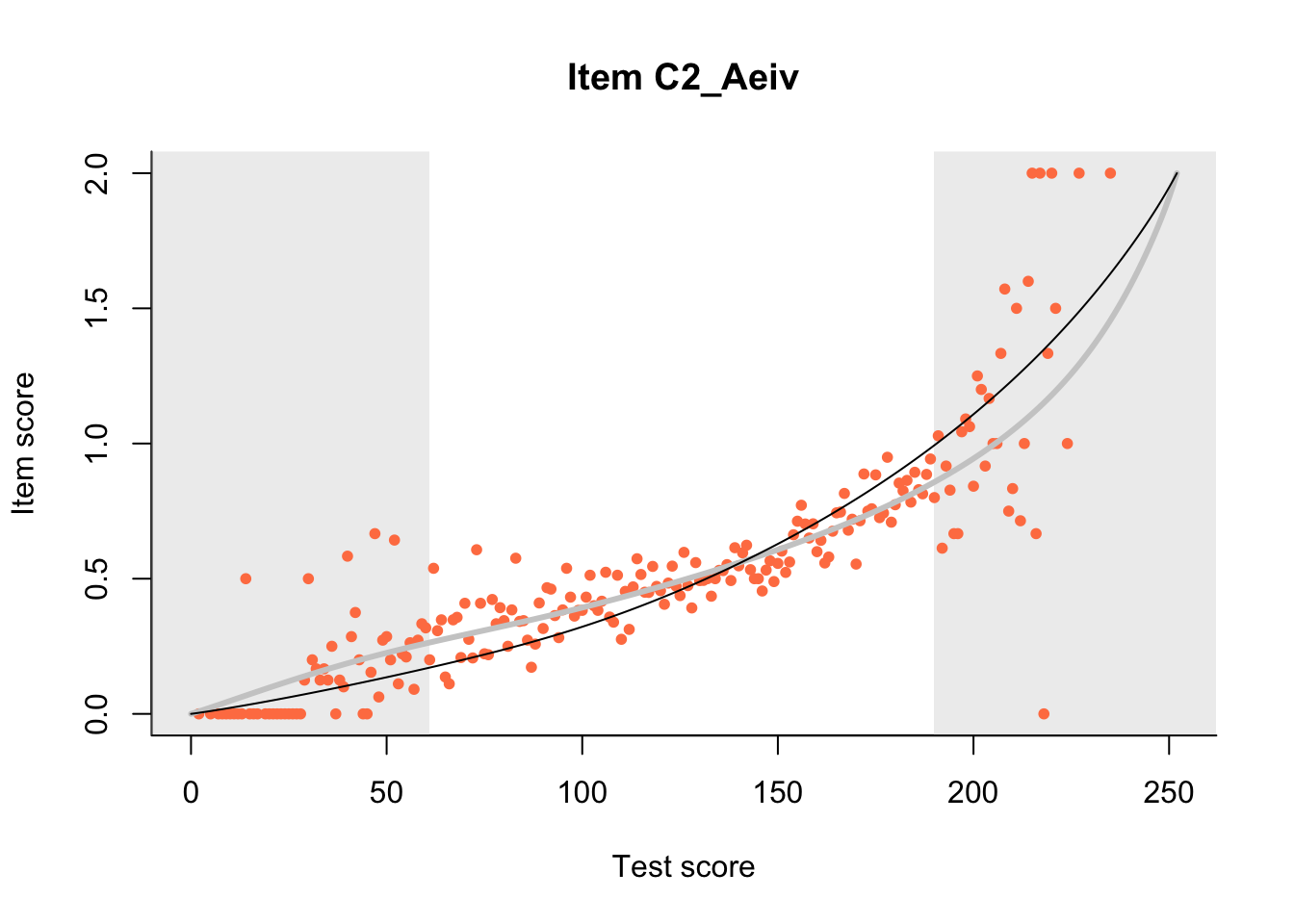
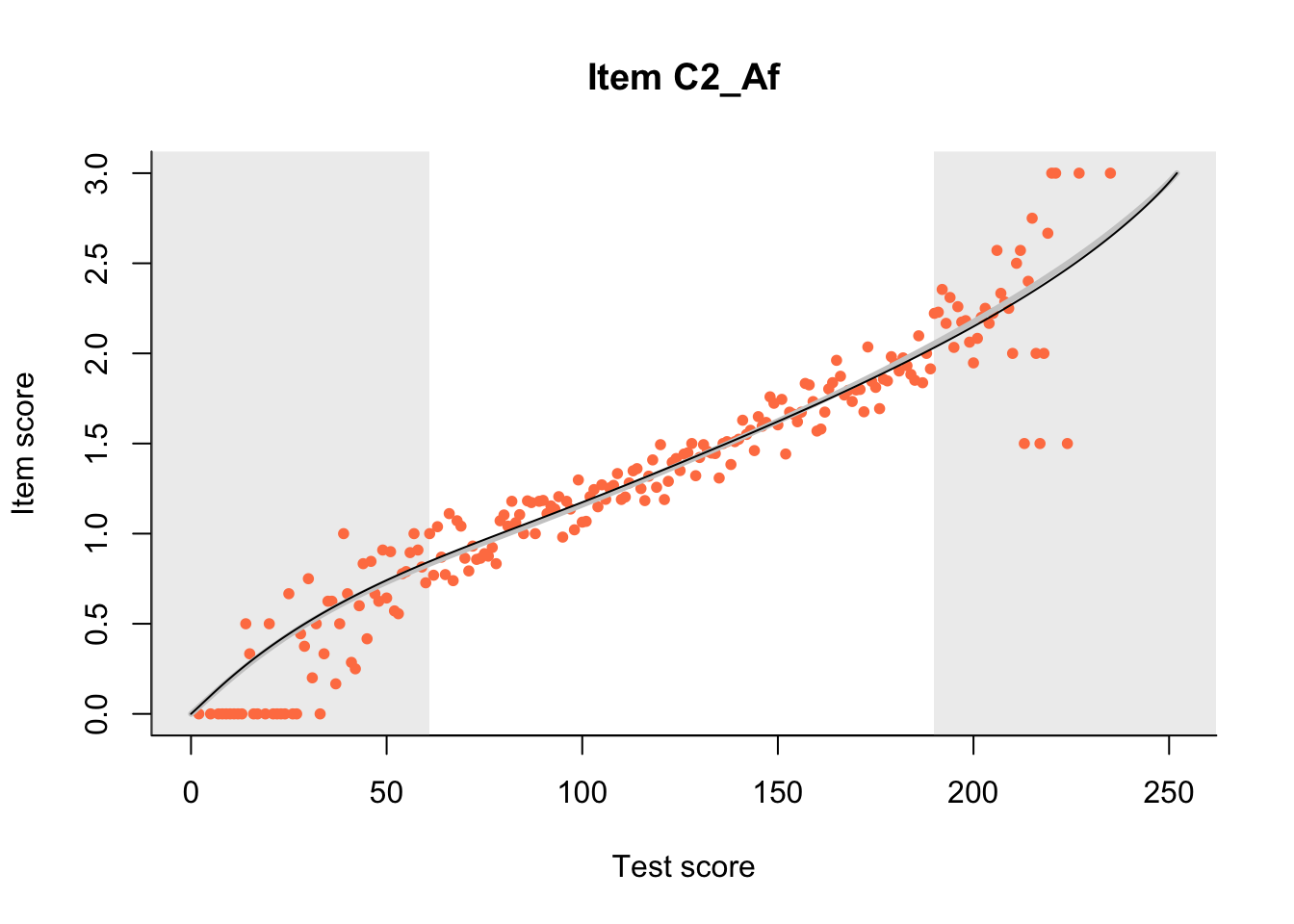
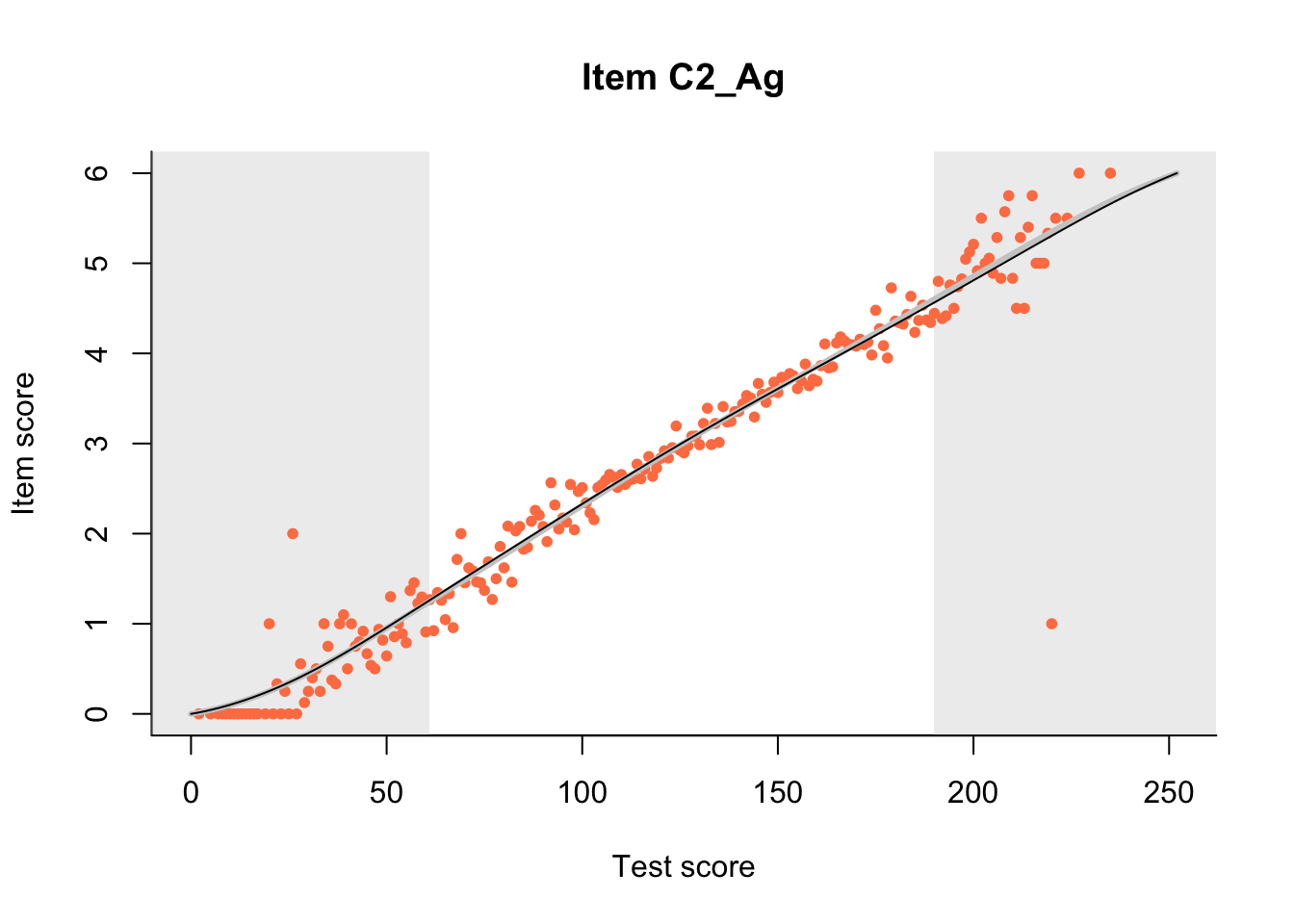
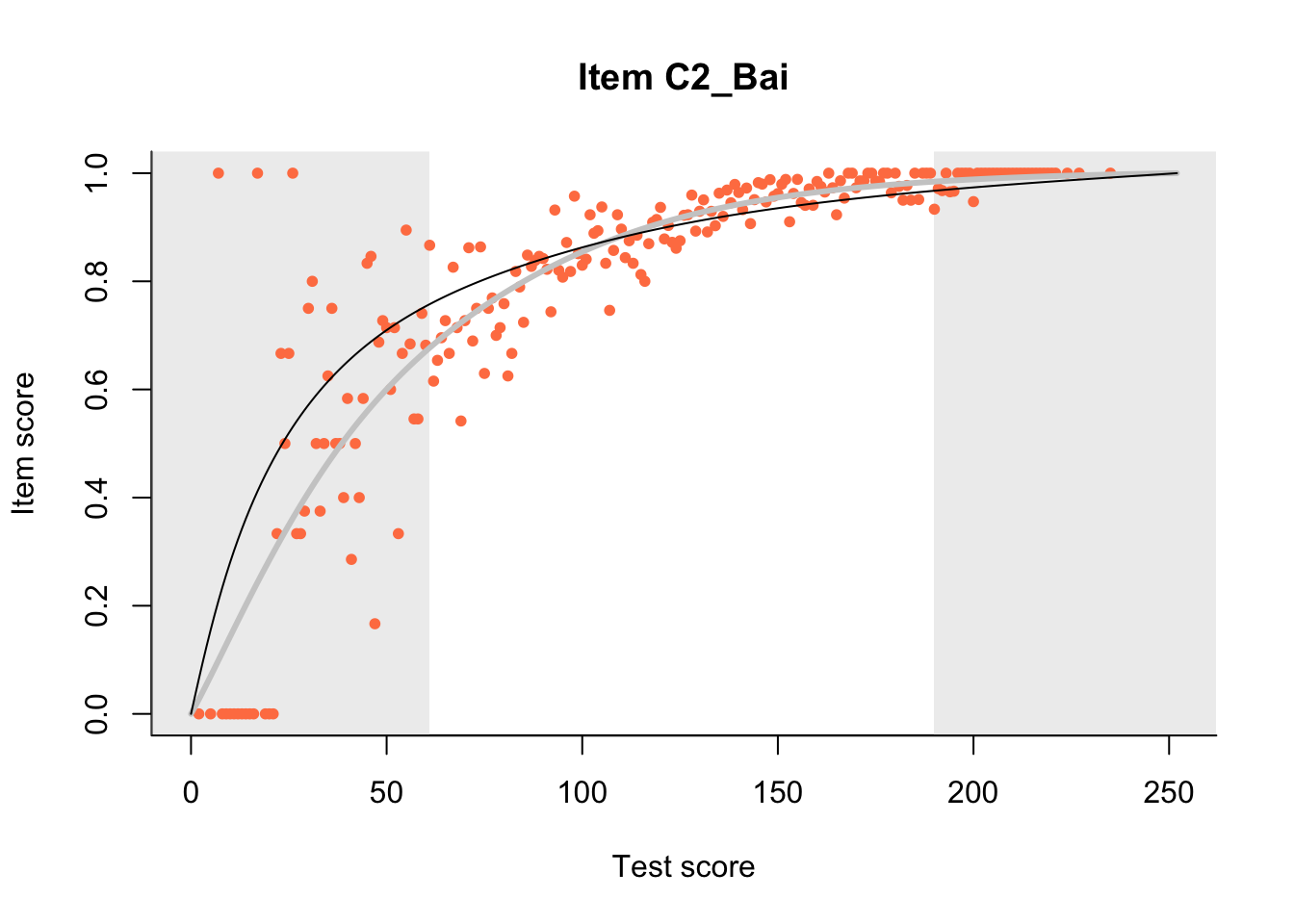
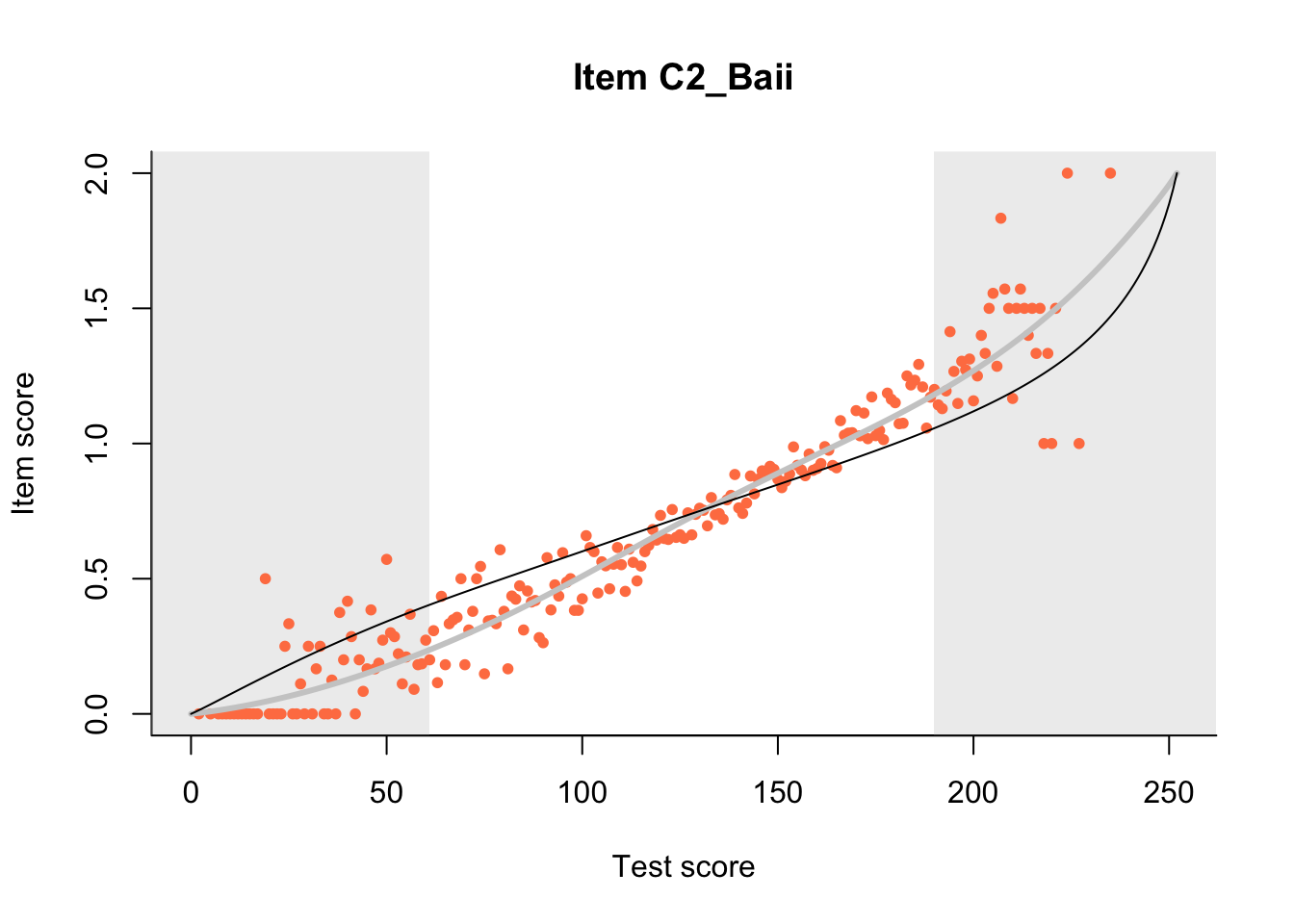
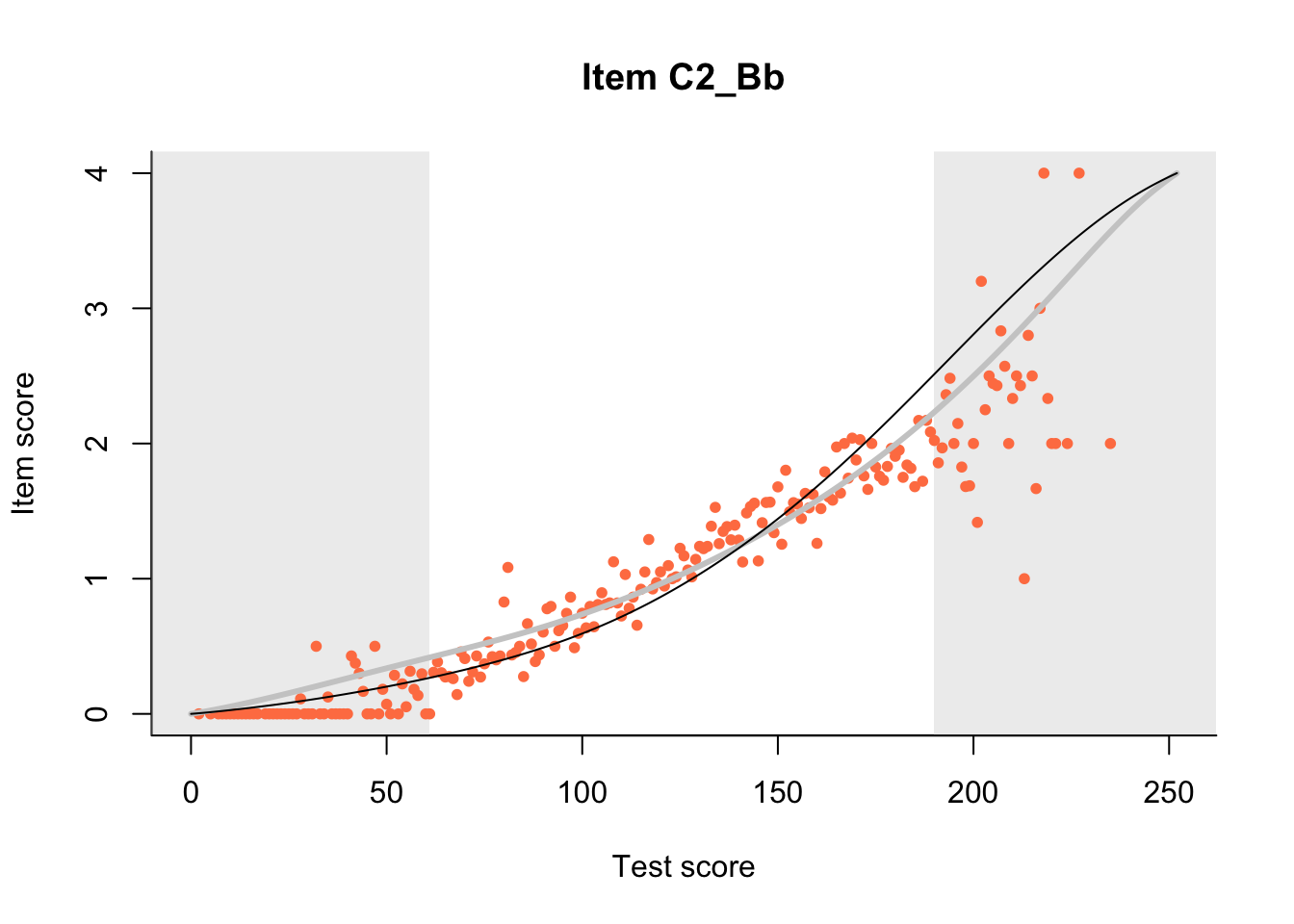
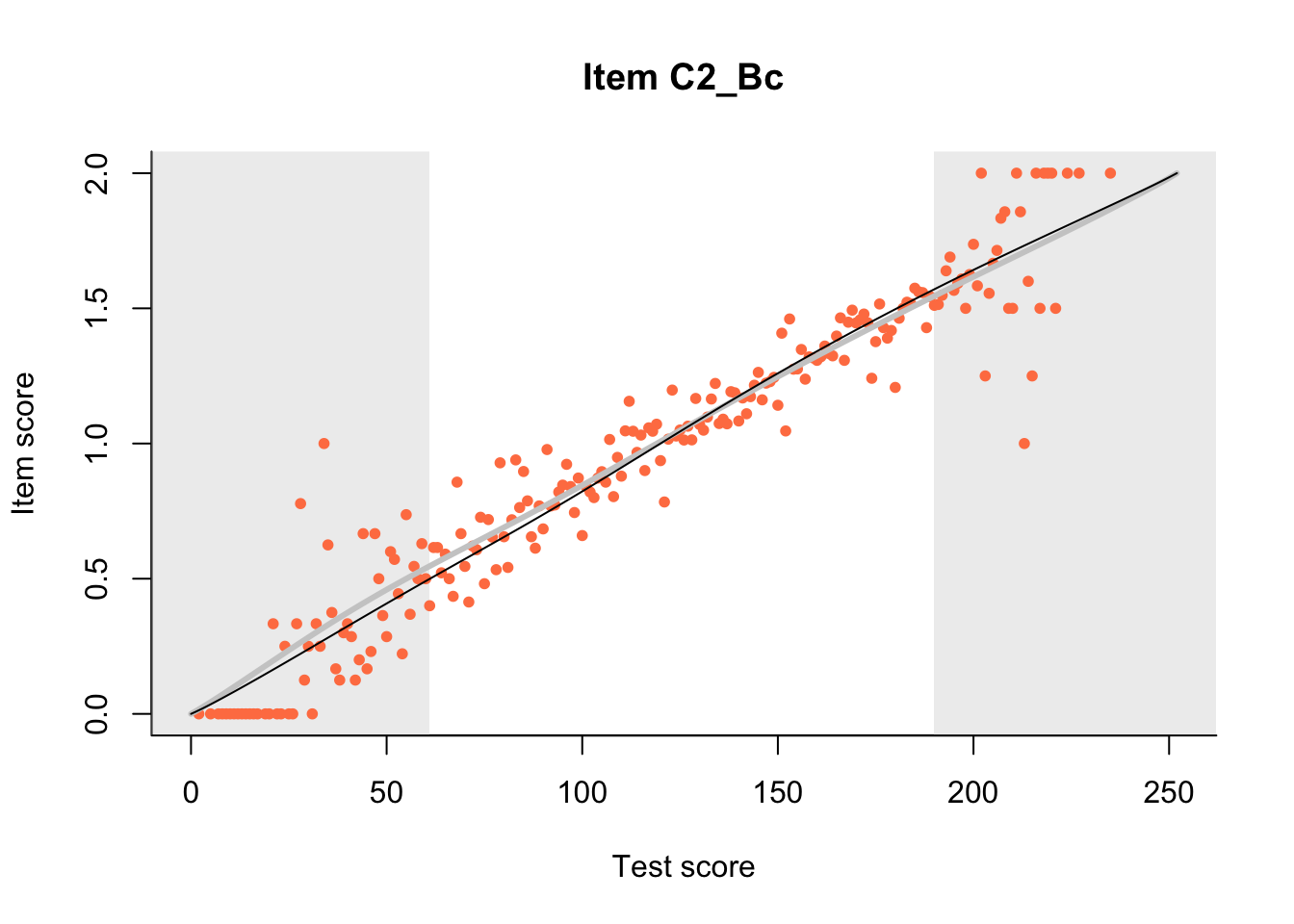
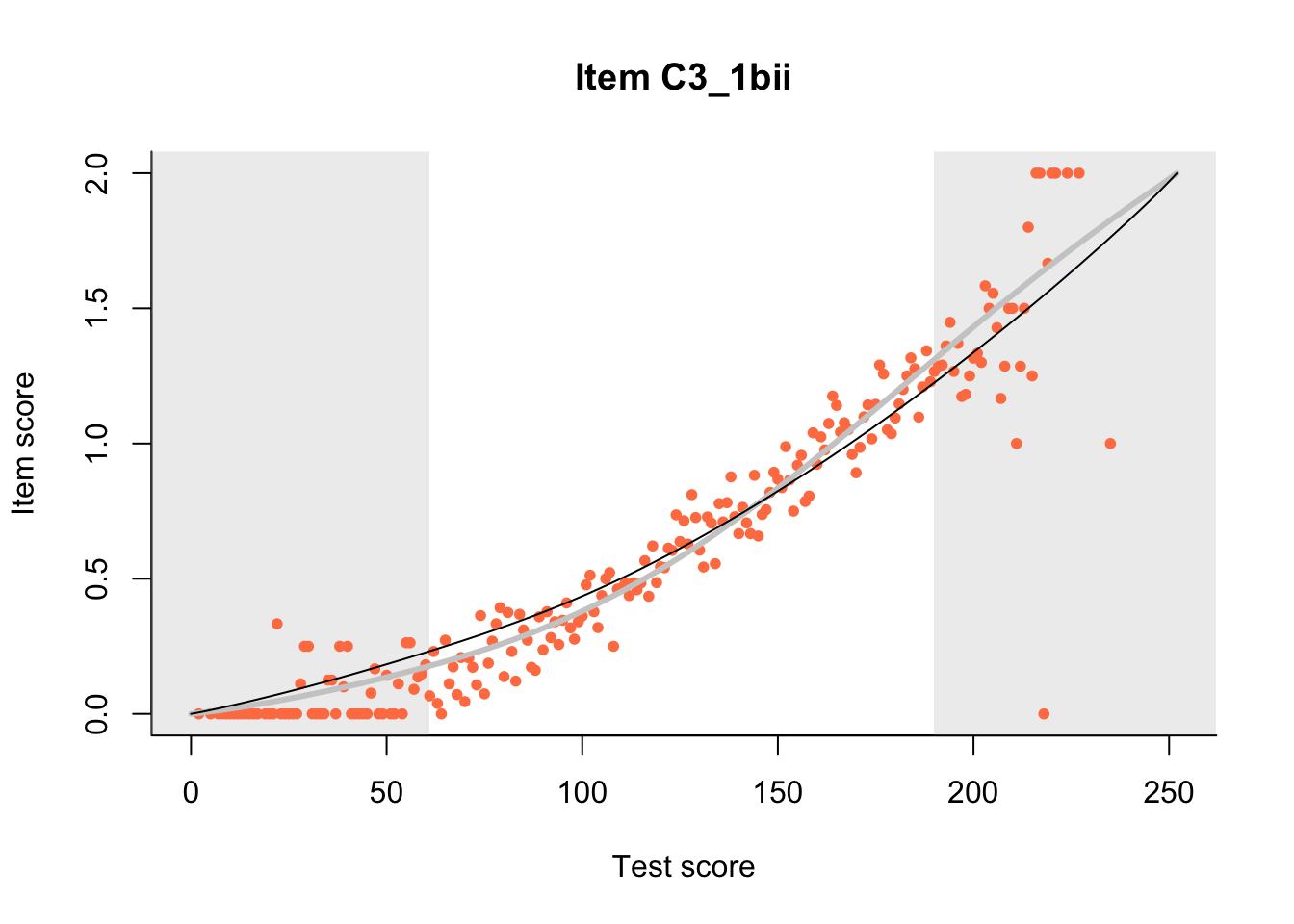
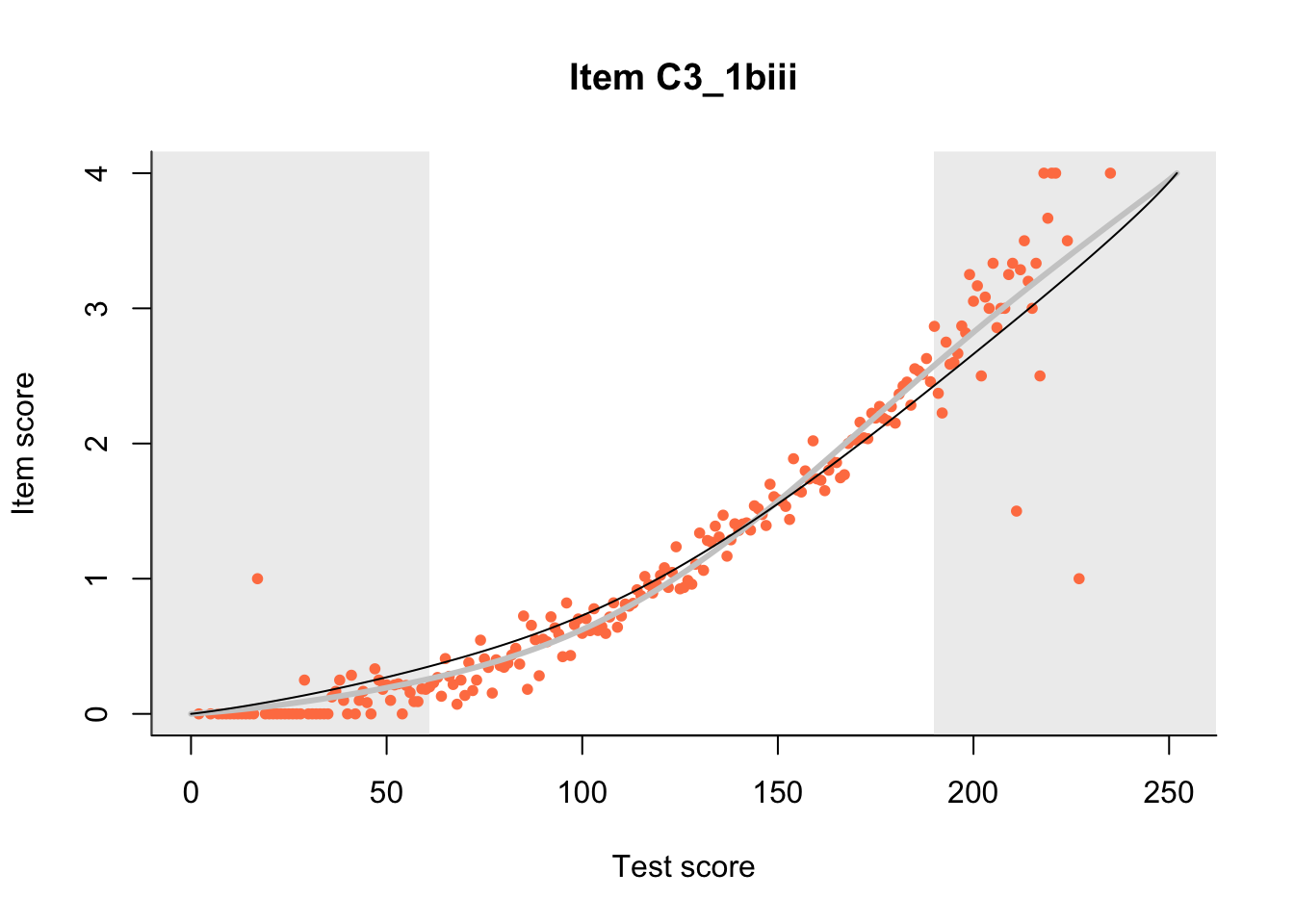
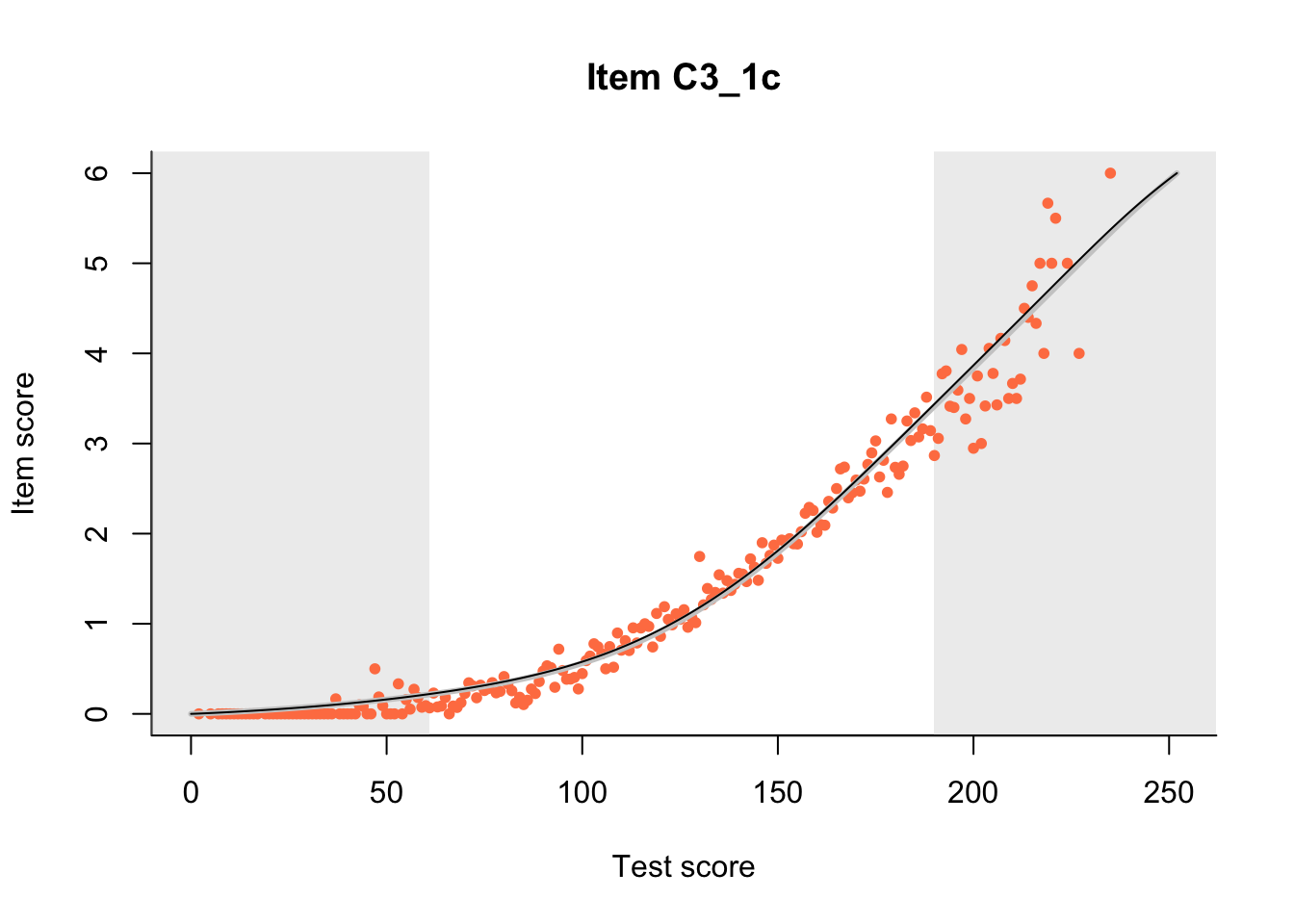
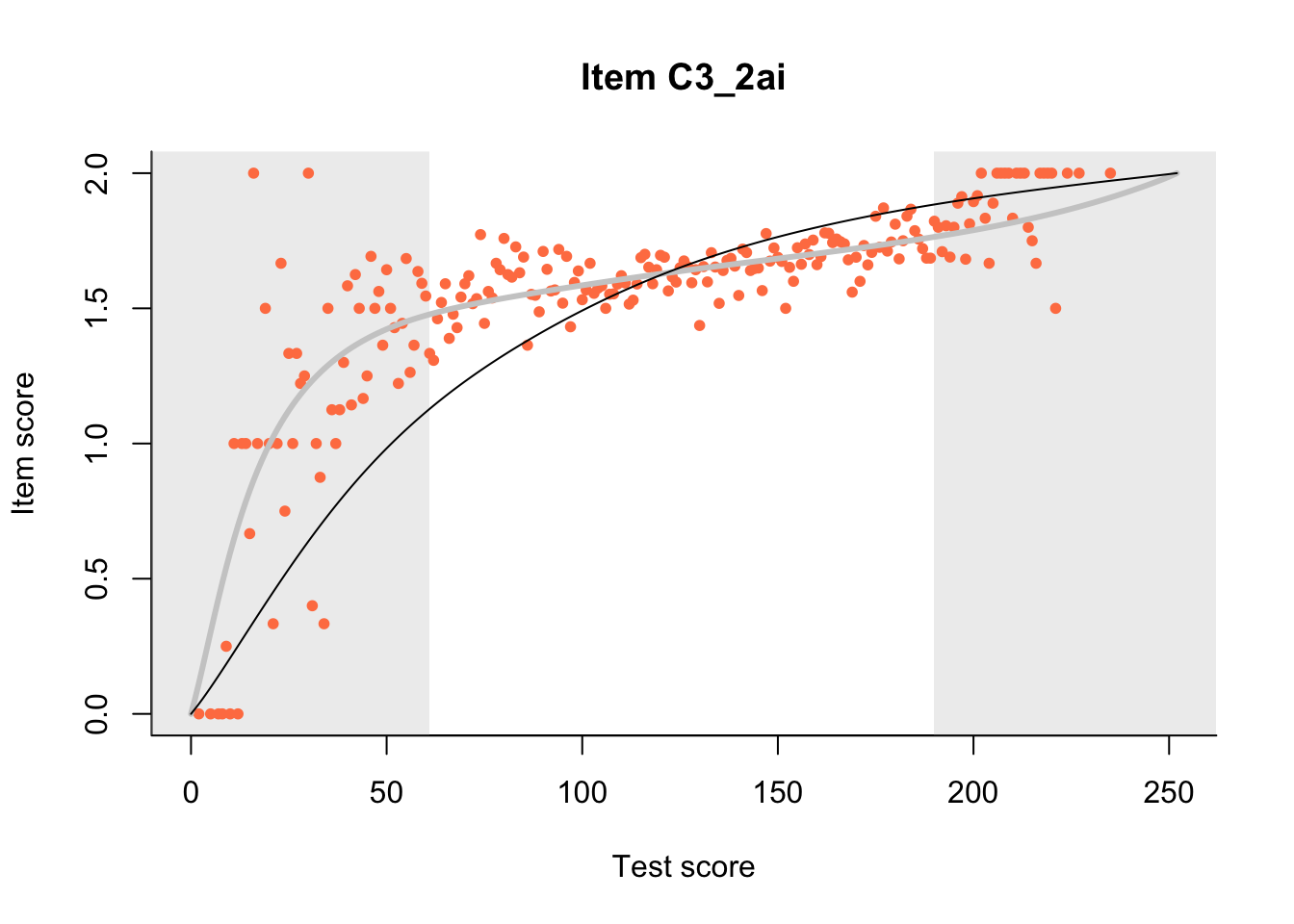
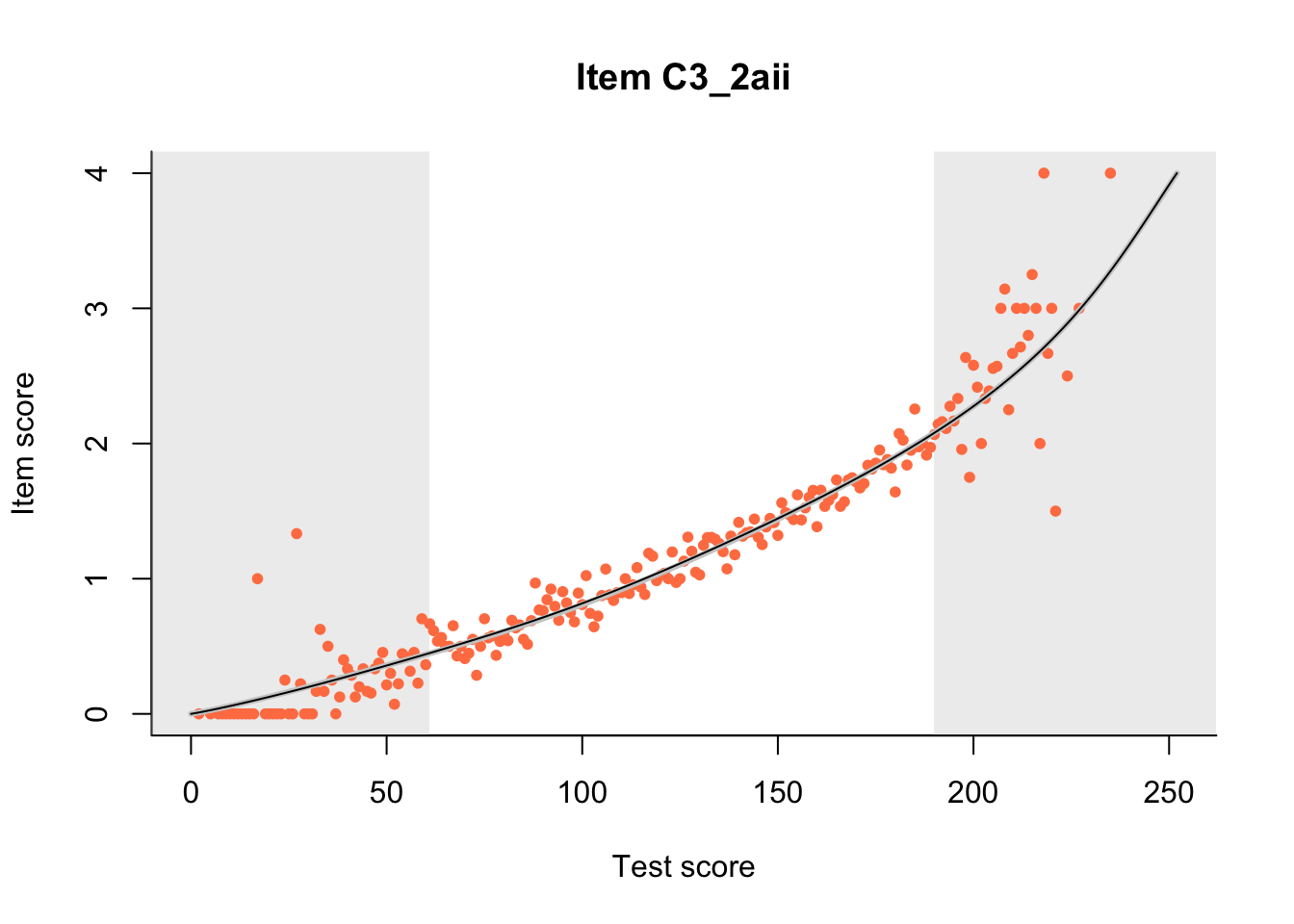
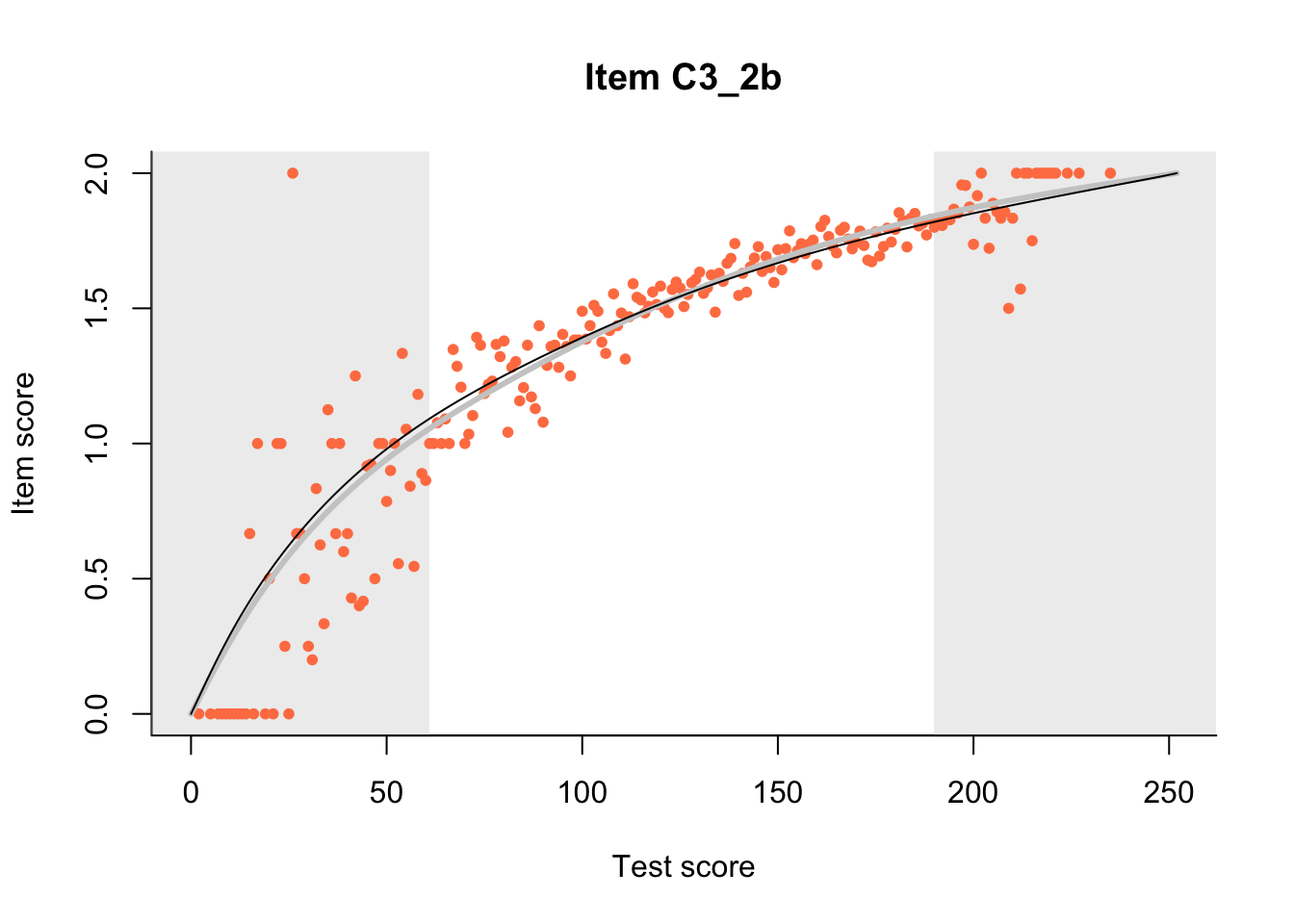
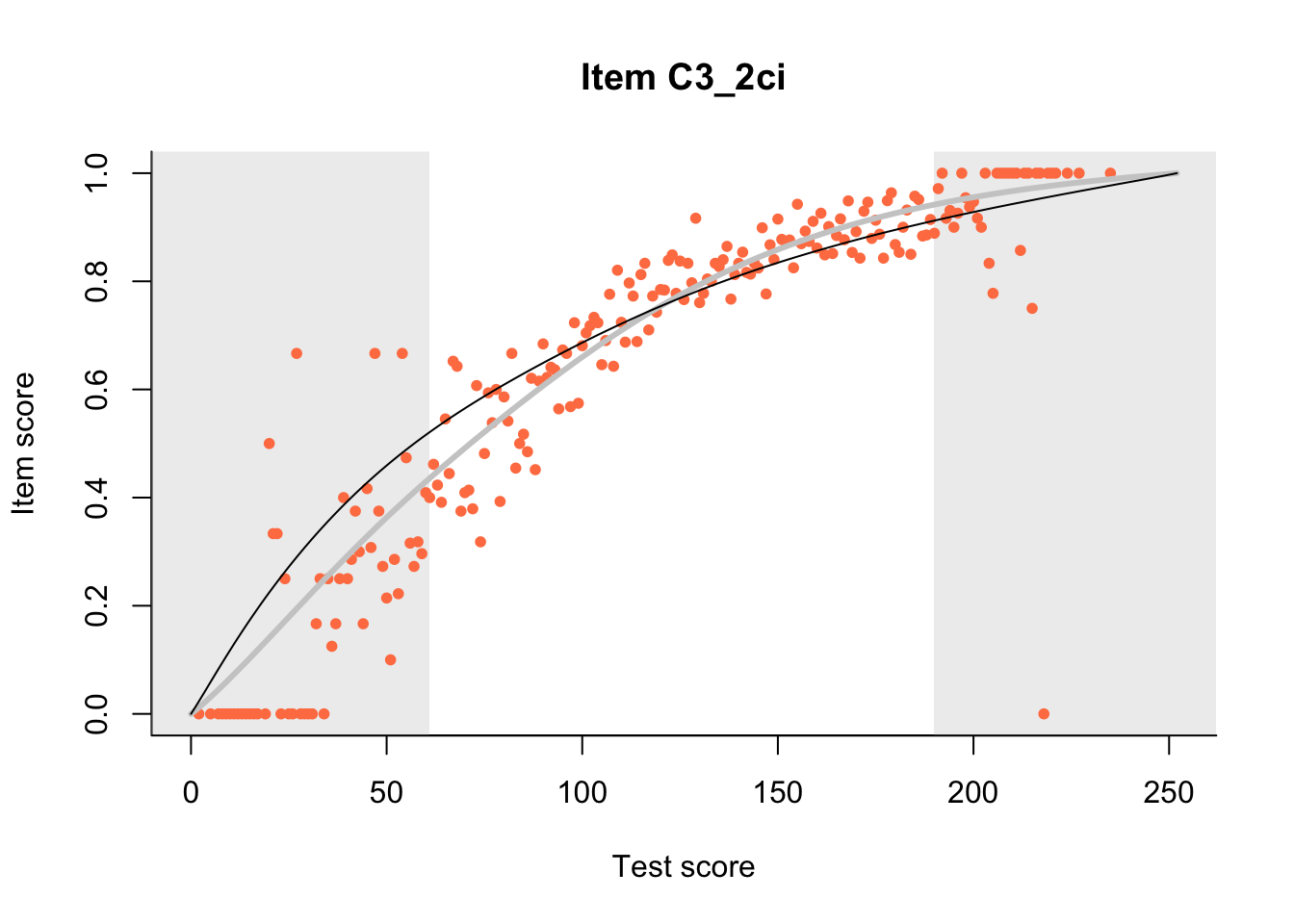
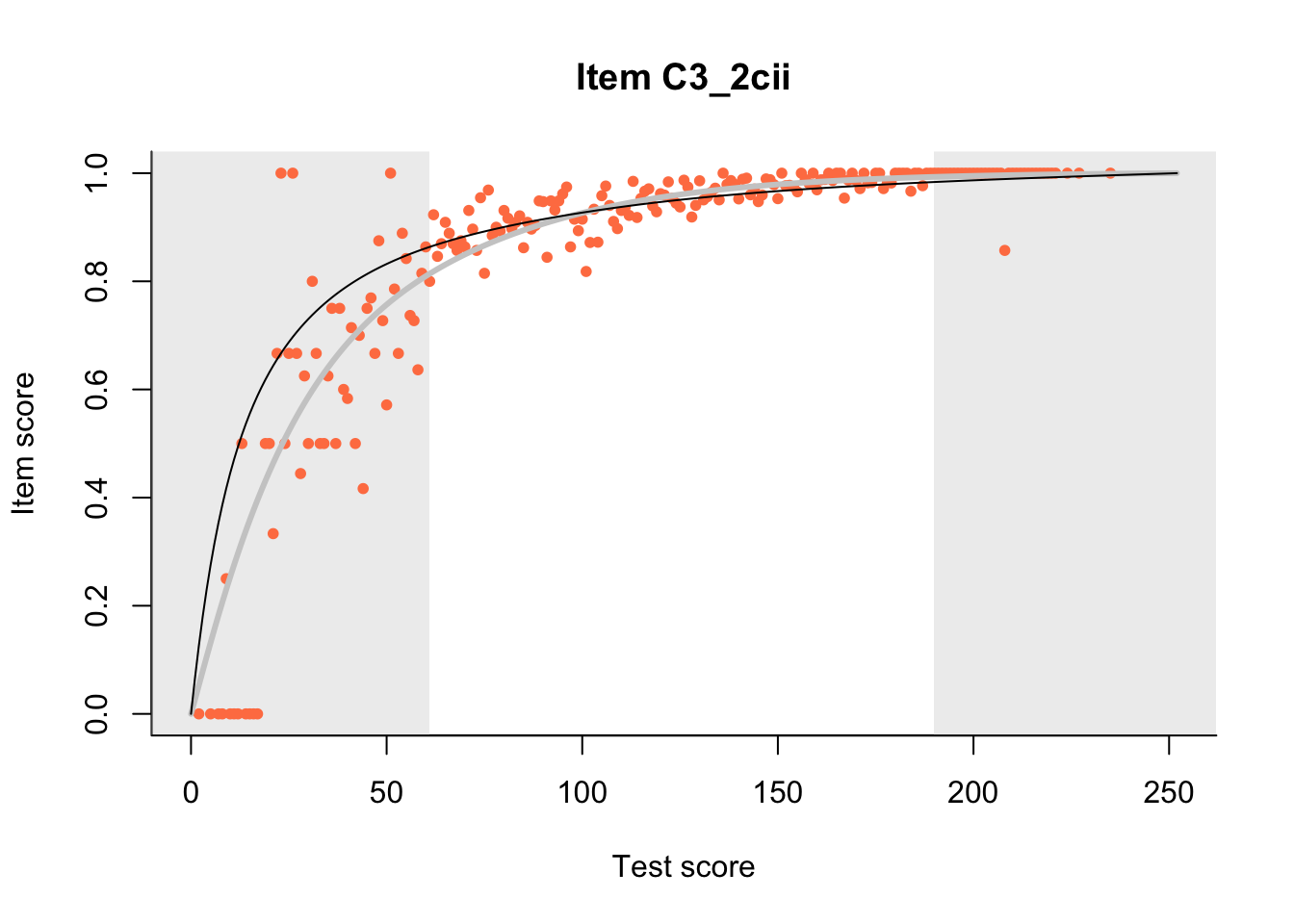
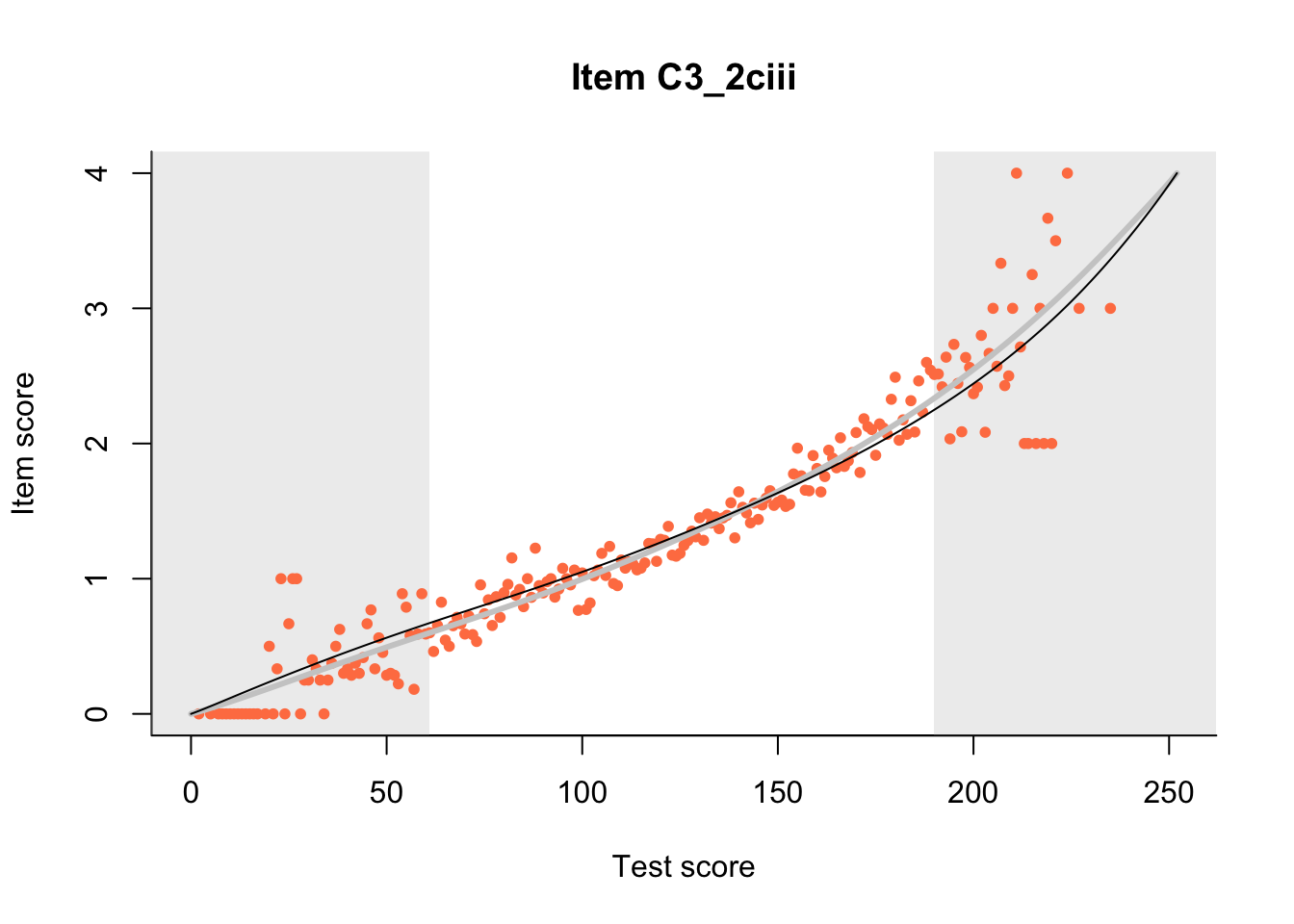
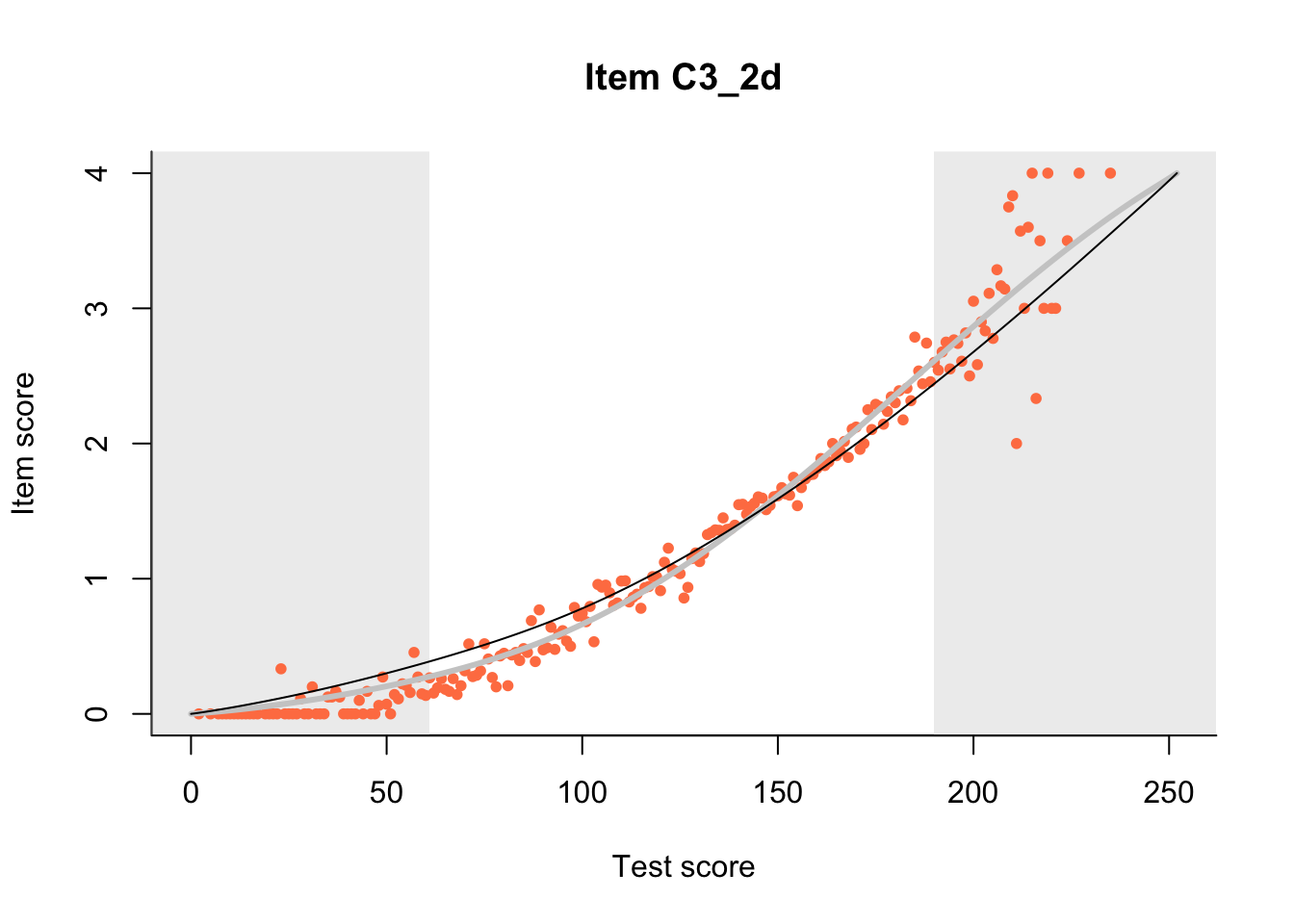
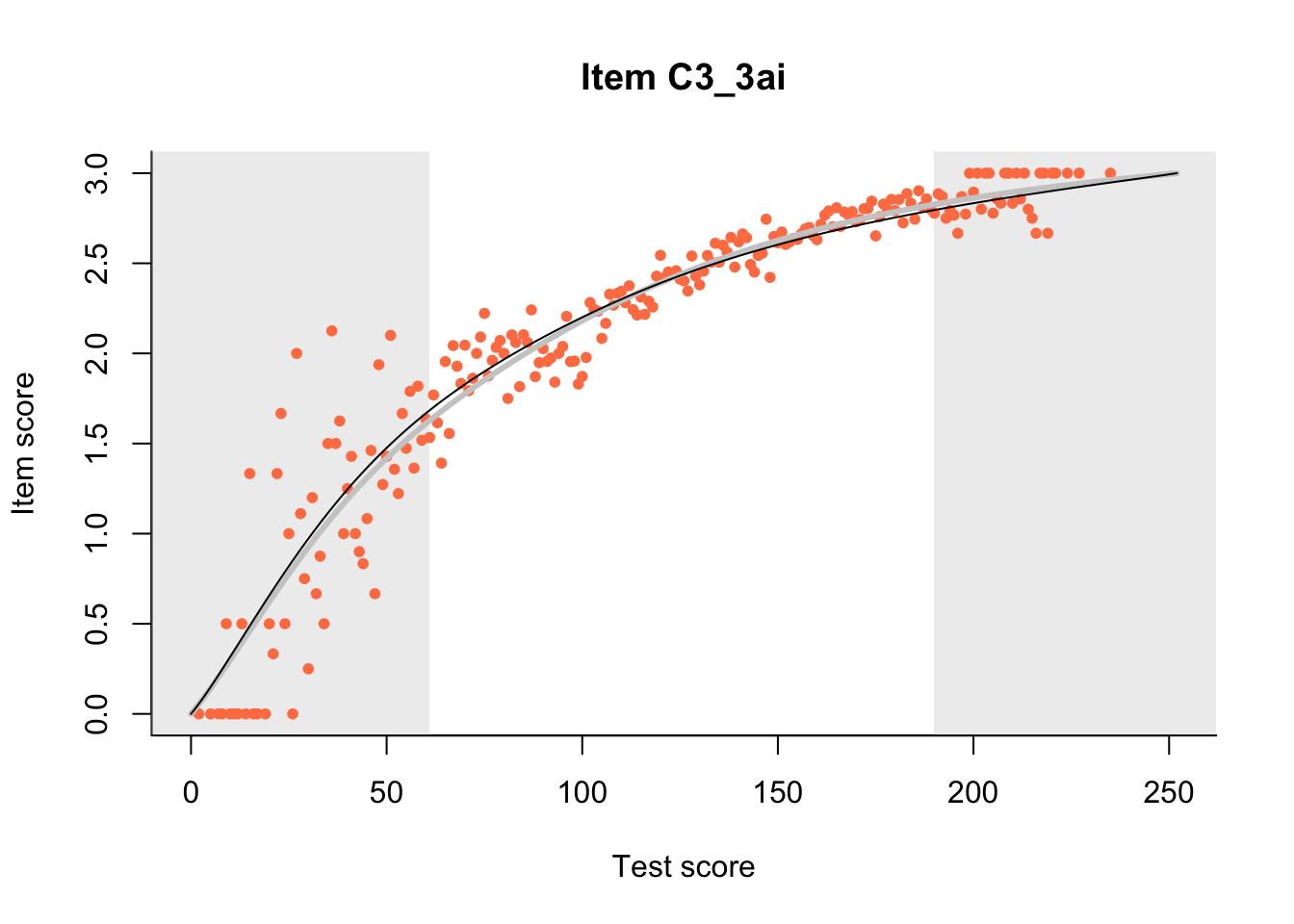
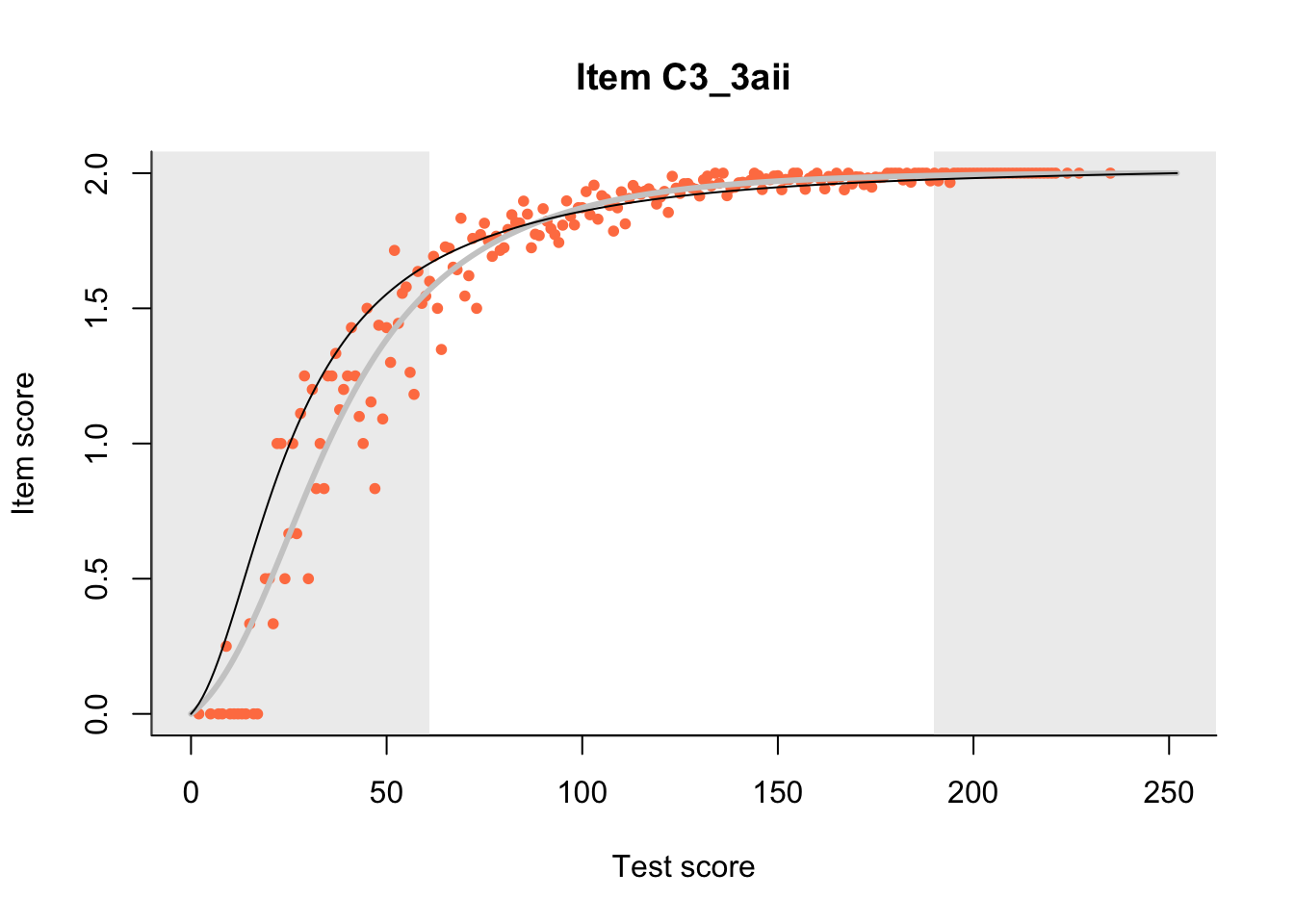
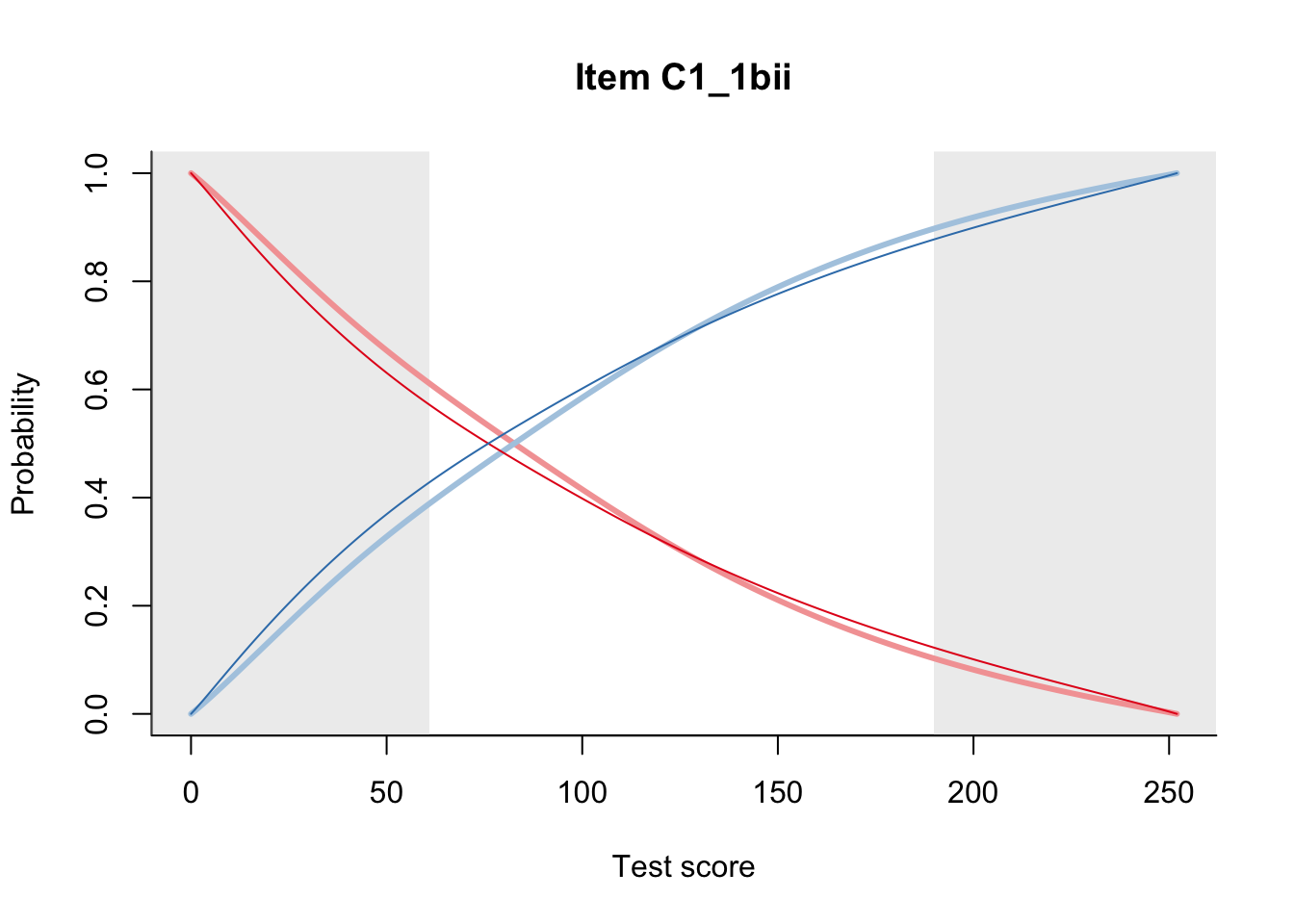
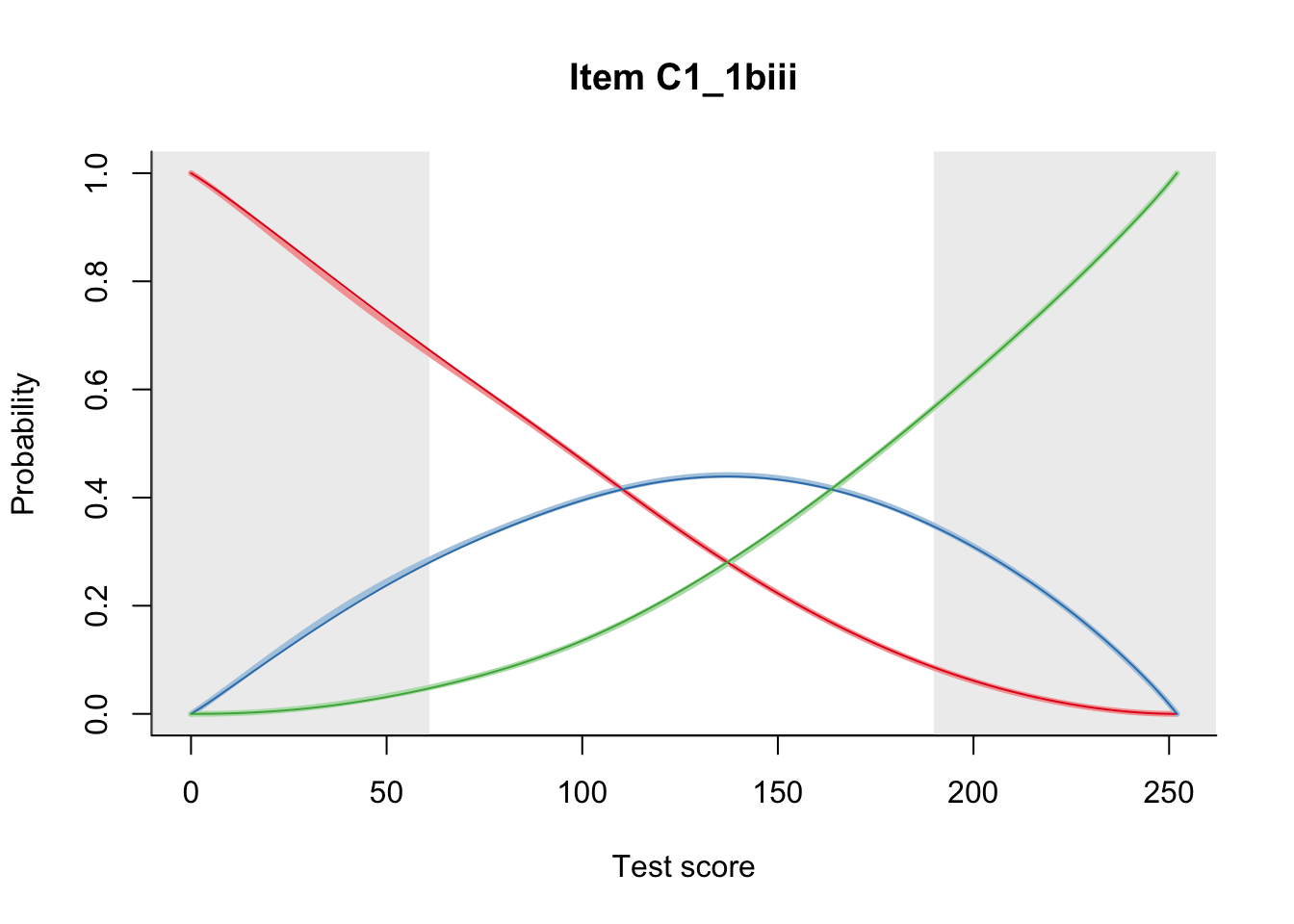
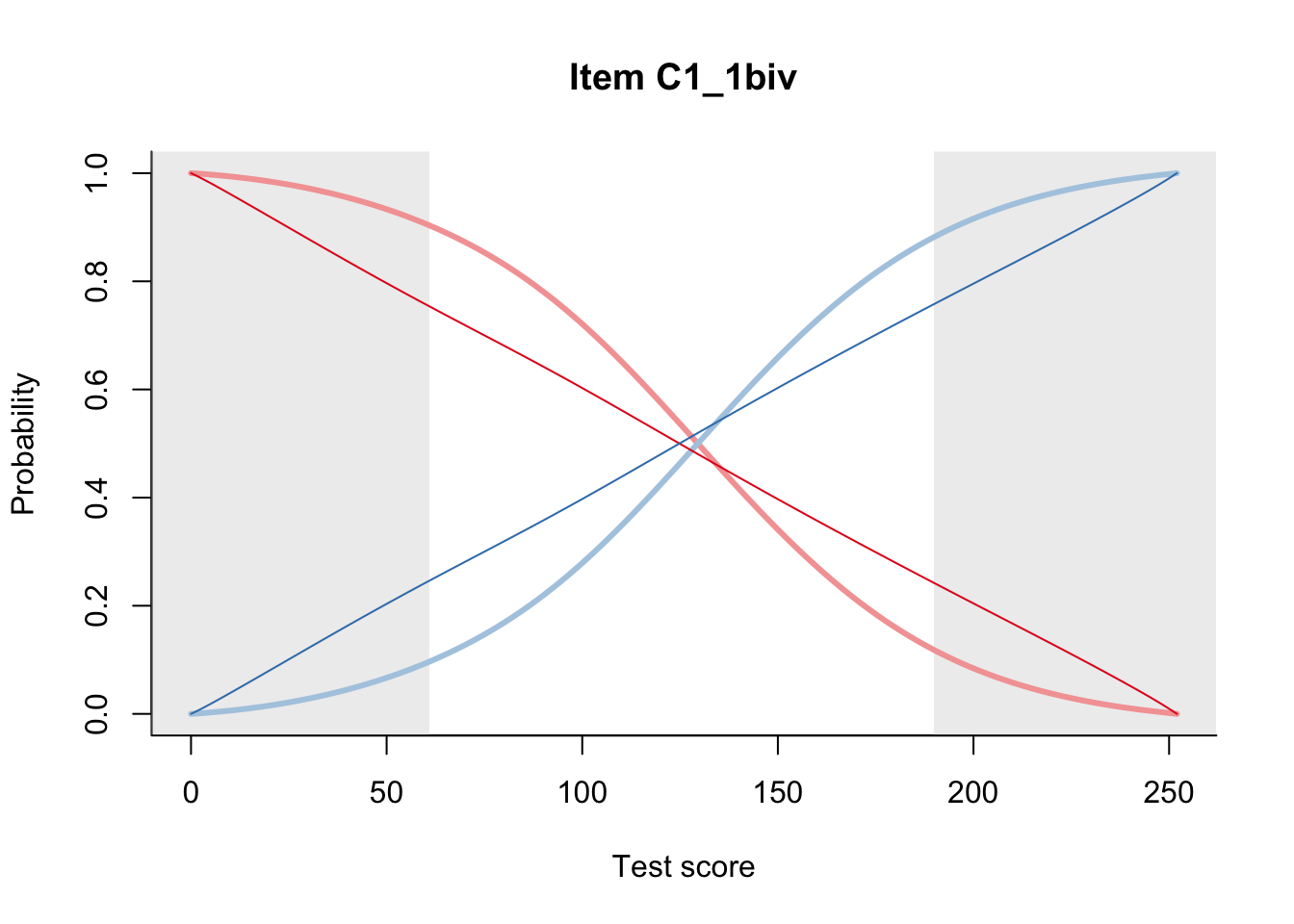
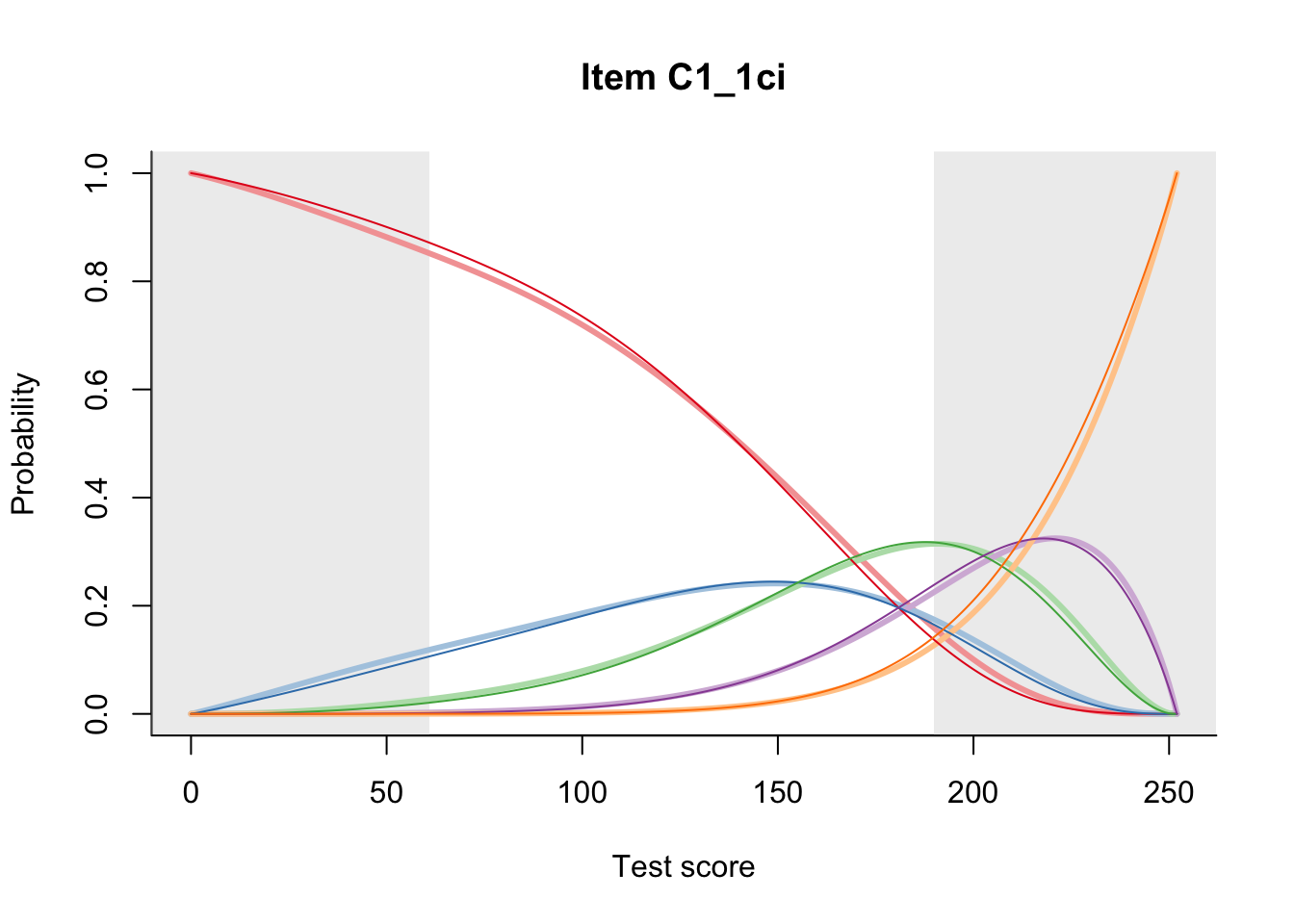
27.1 Expected Score Curves
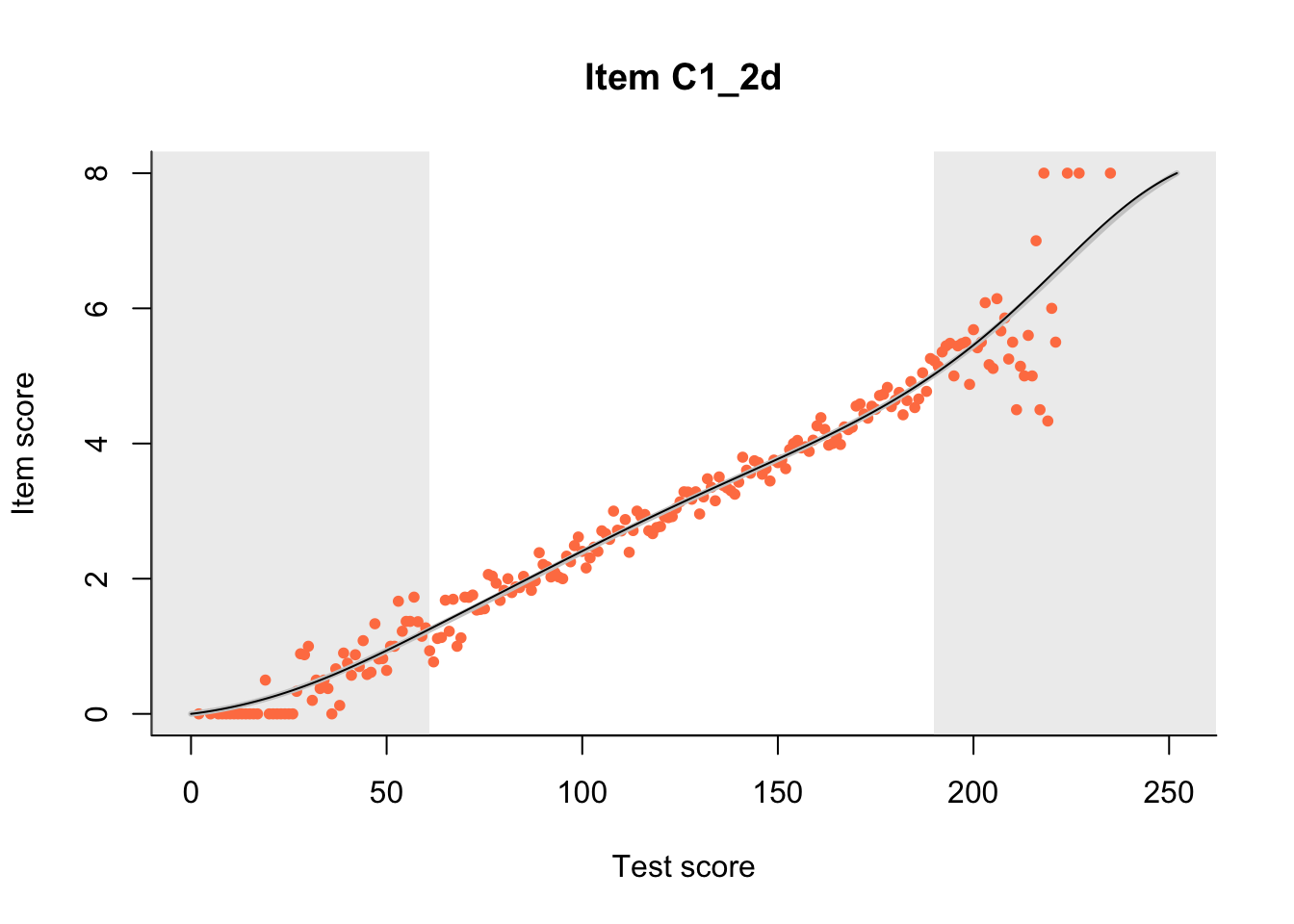
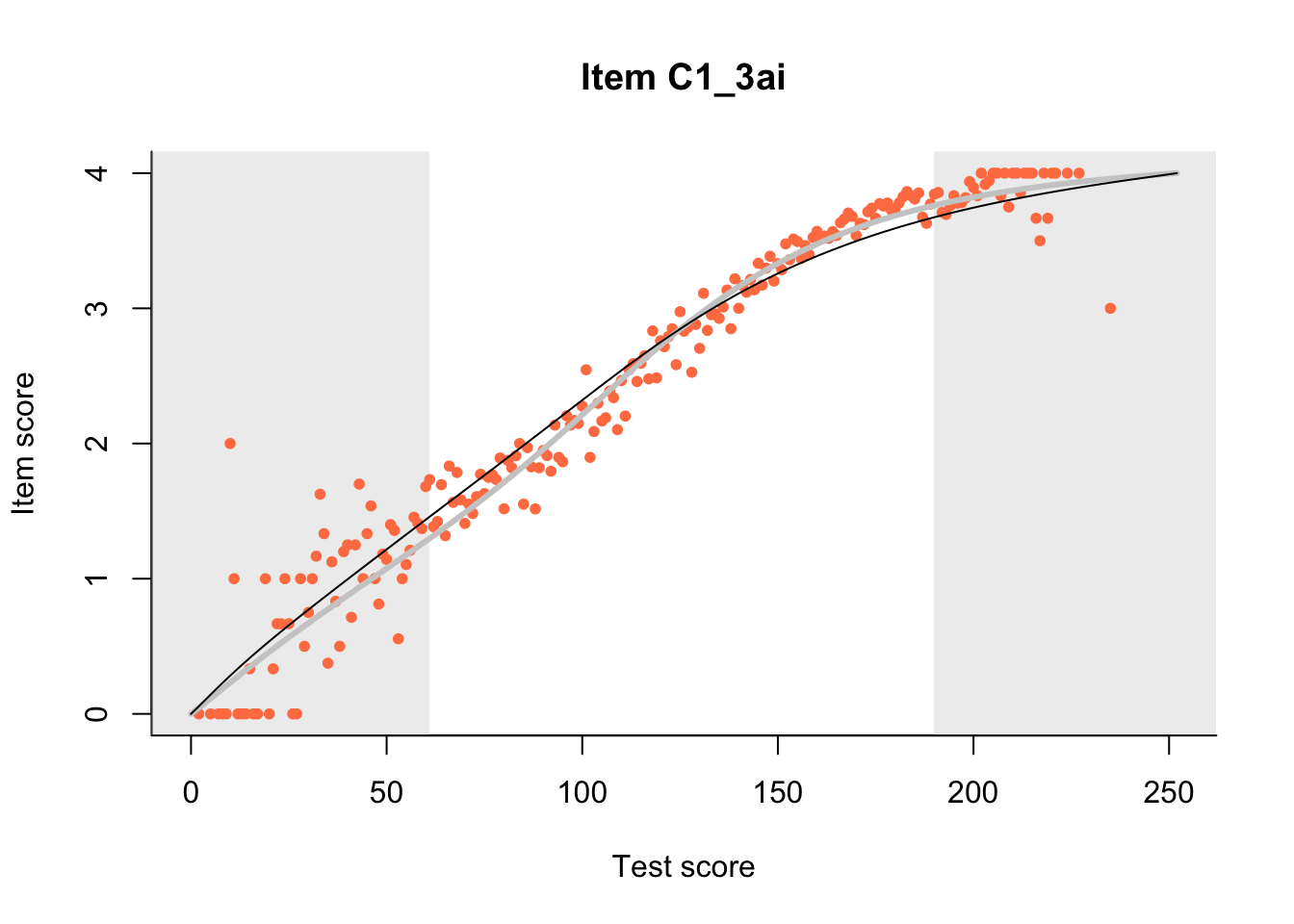
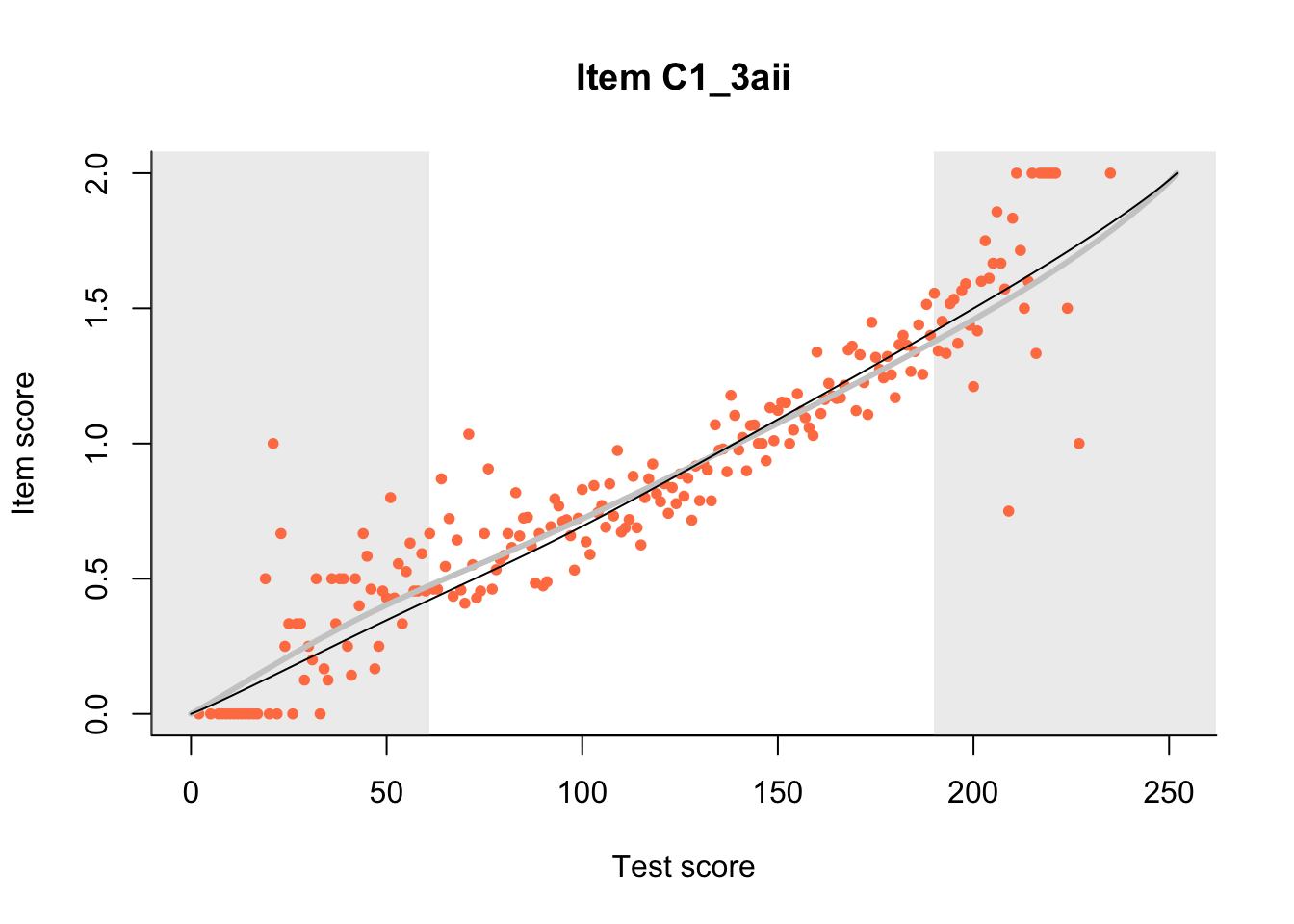
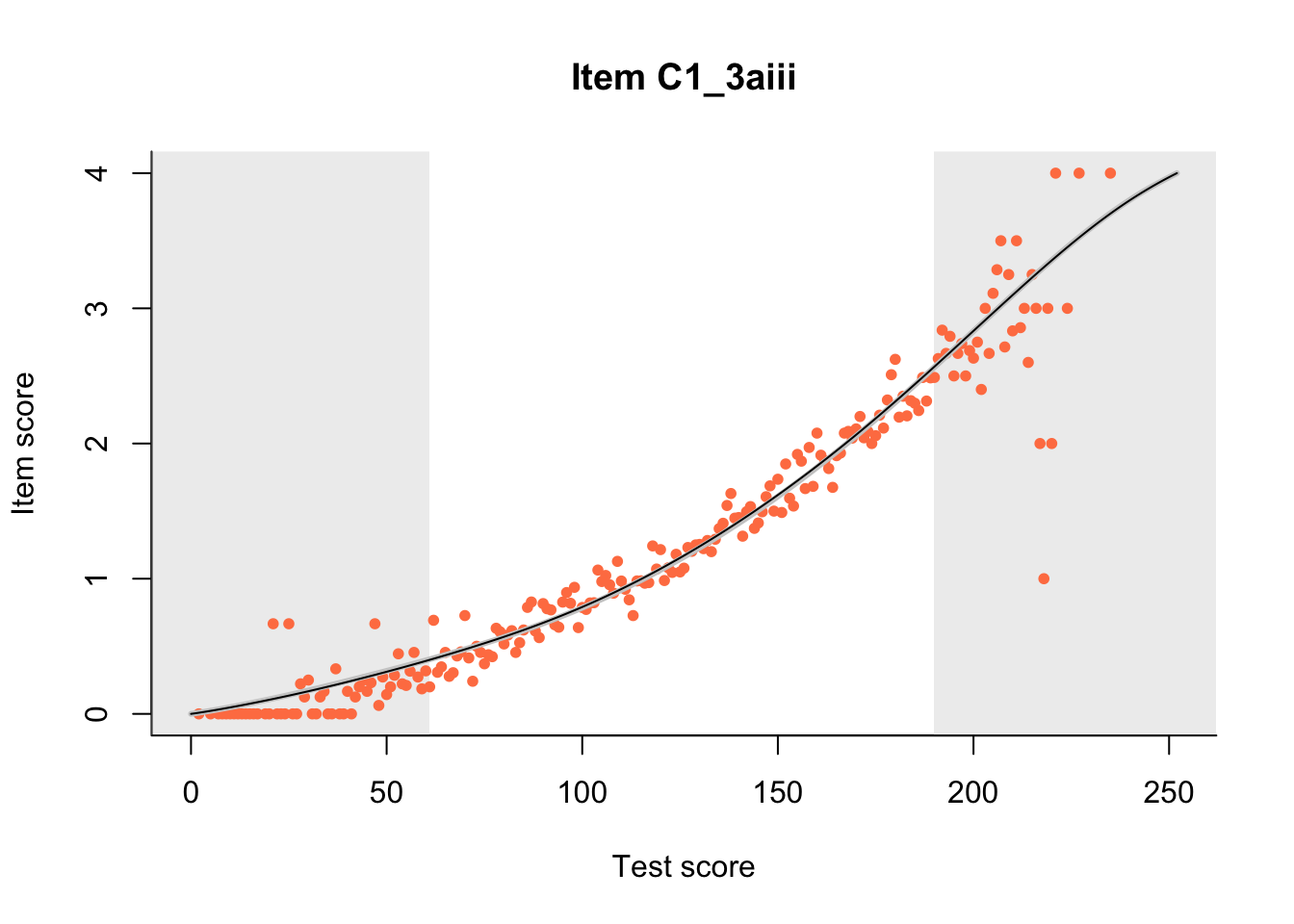
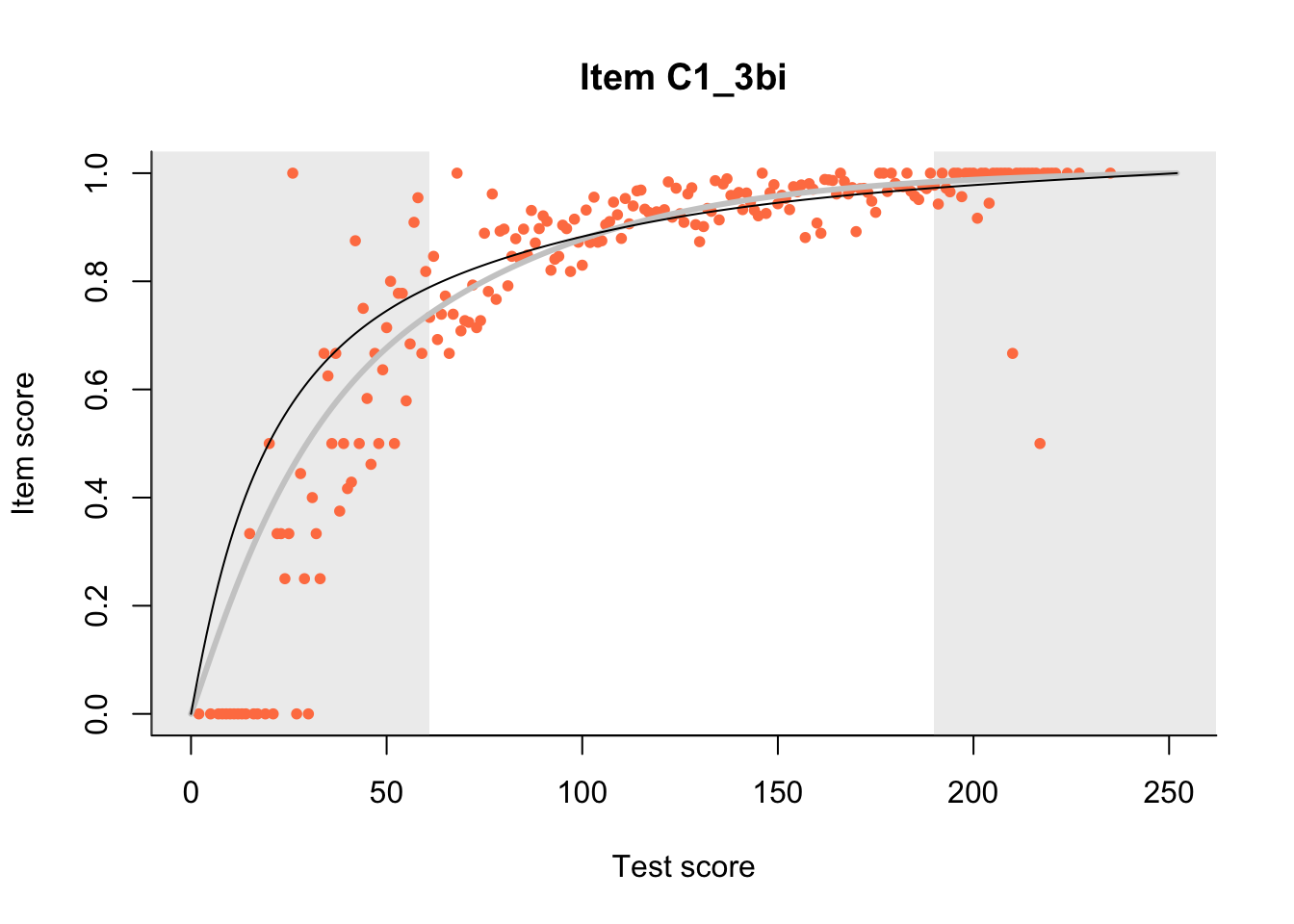
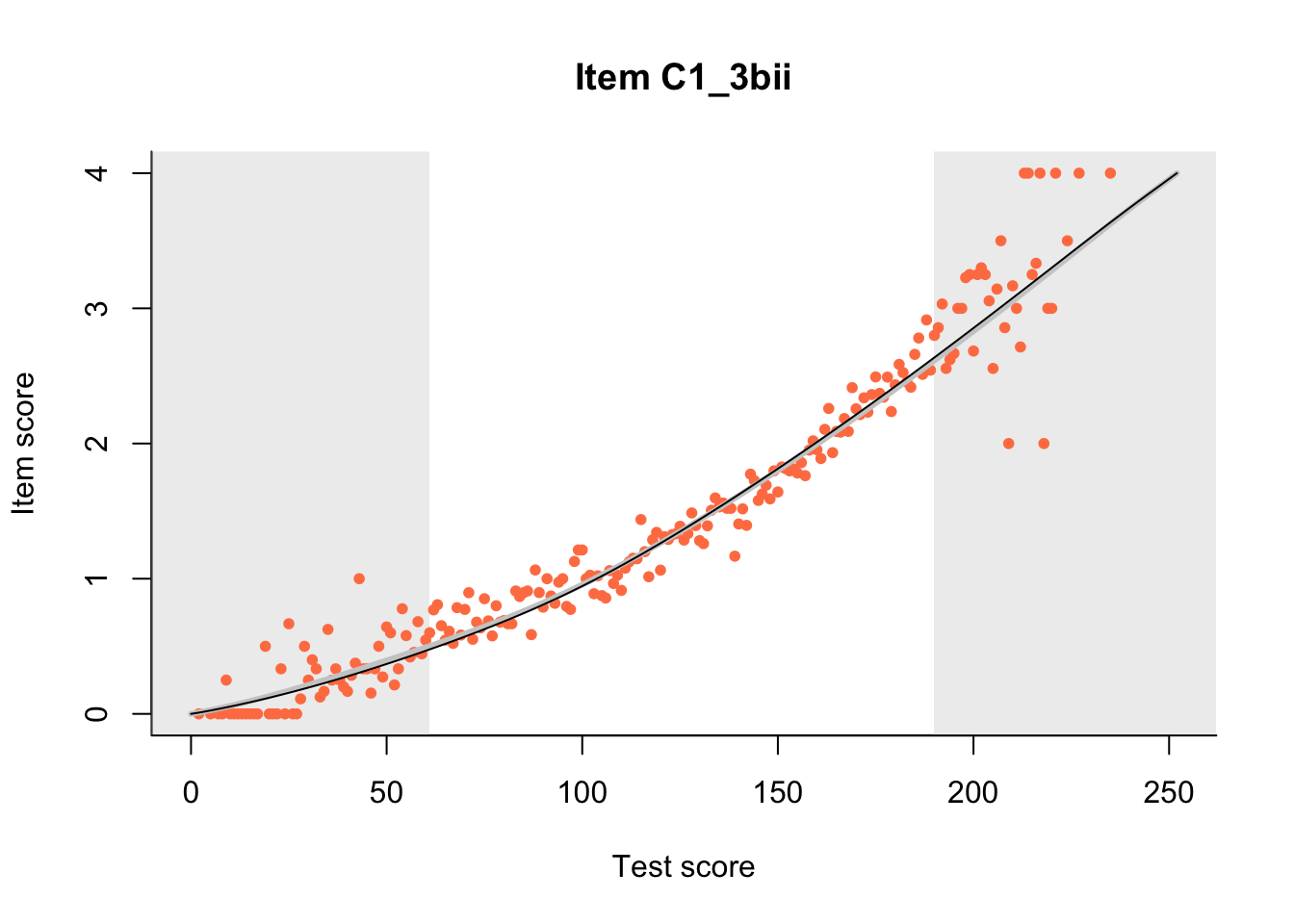
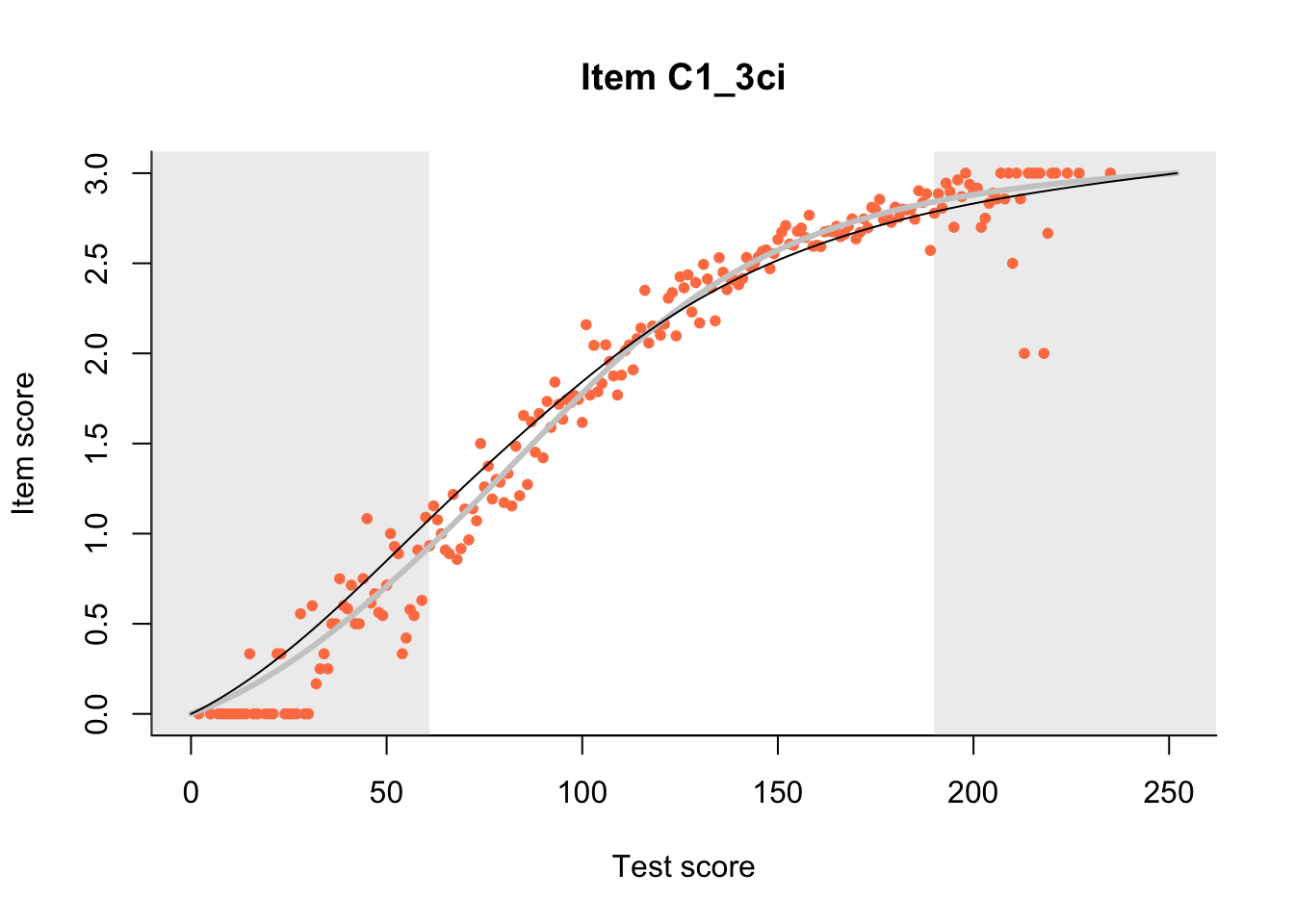
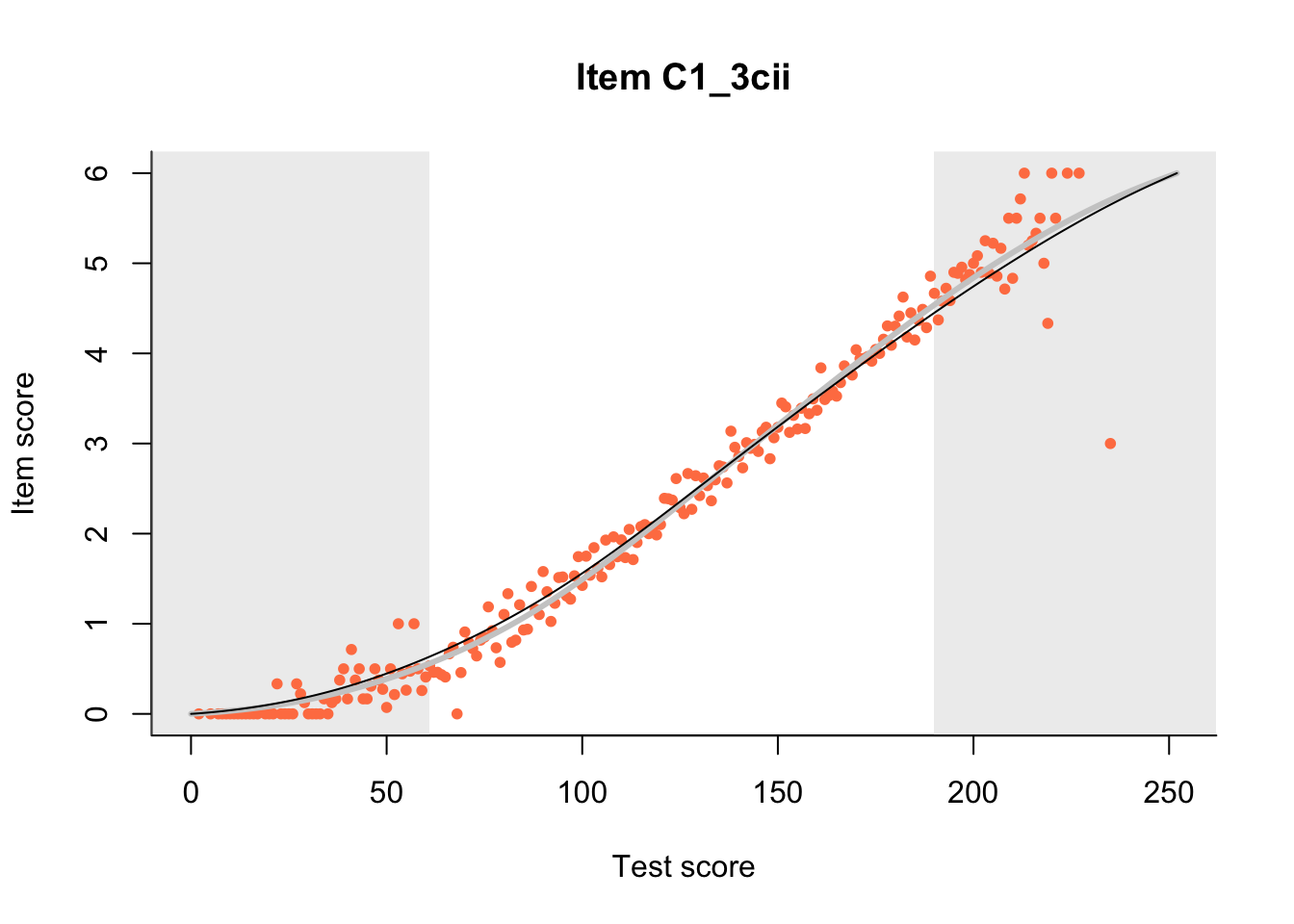
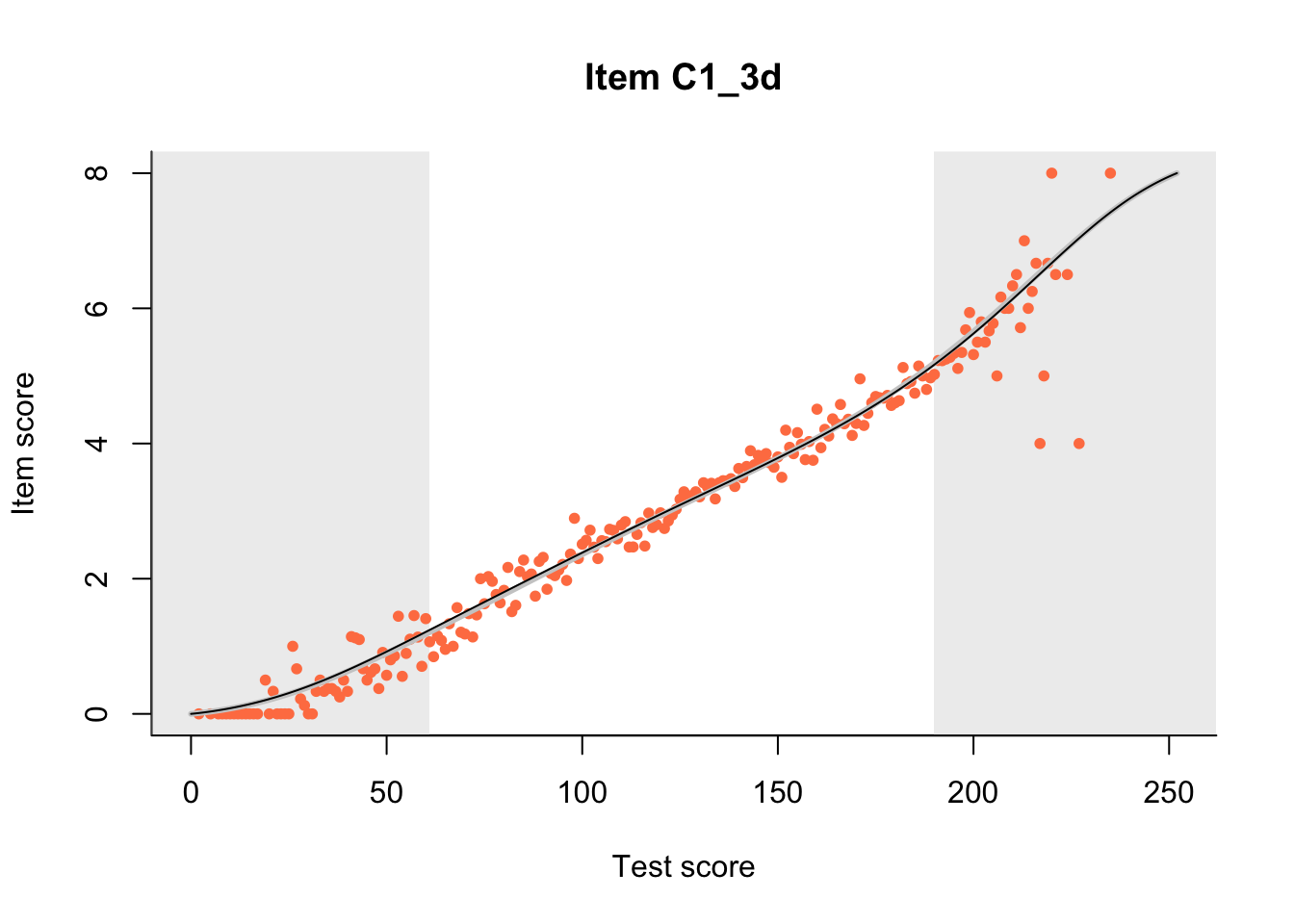
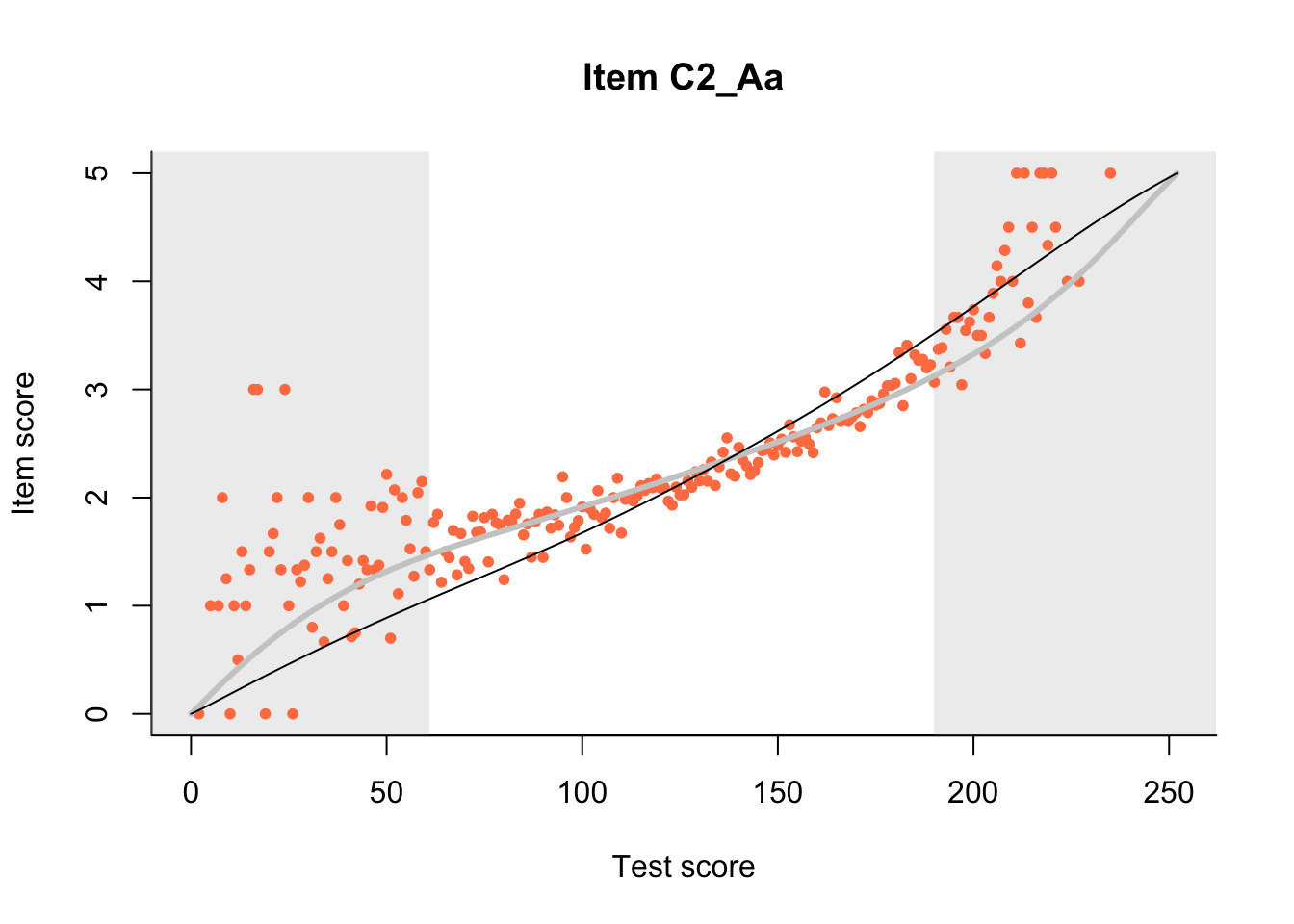
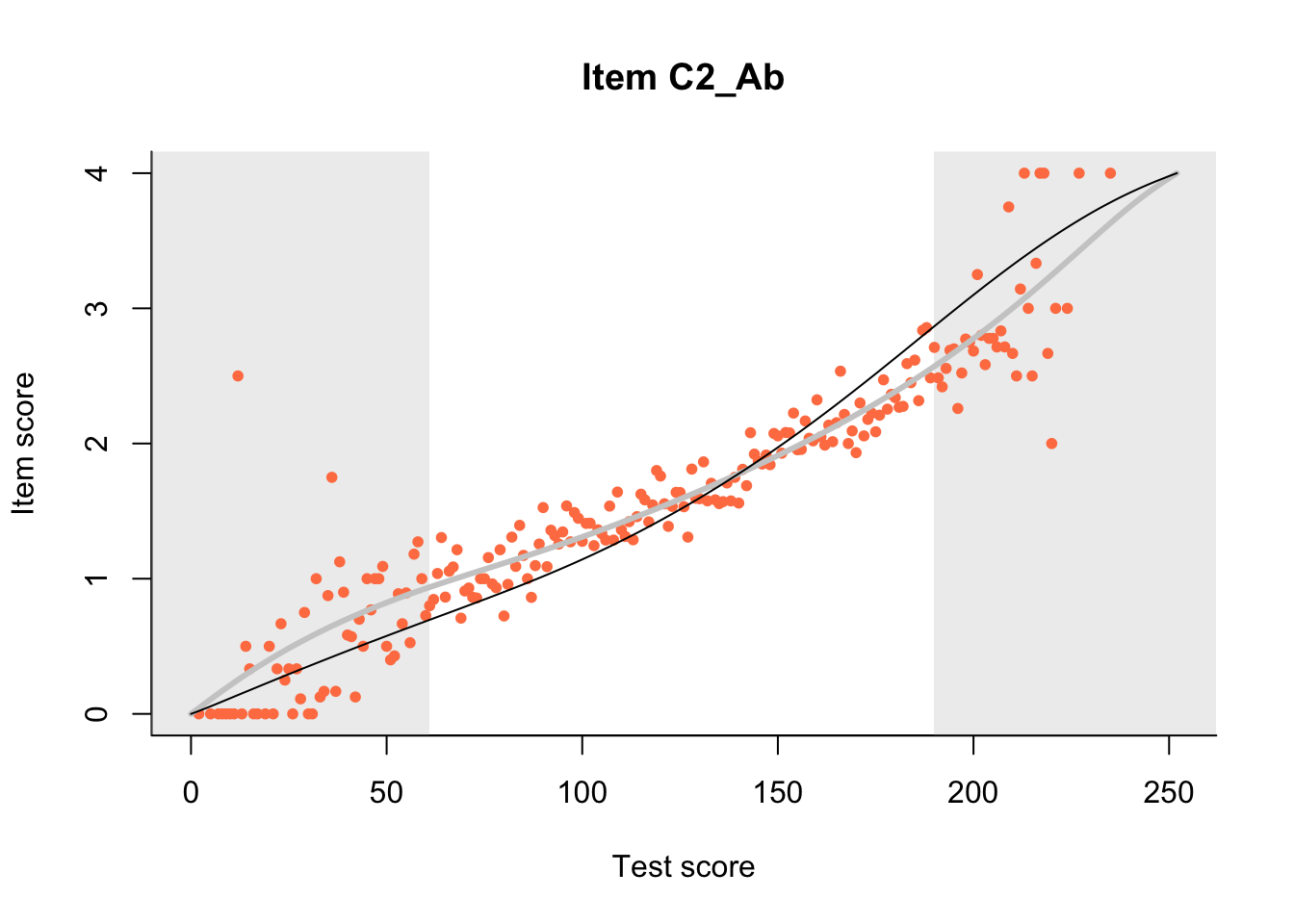
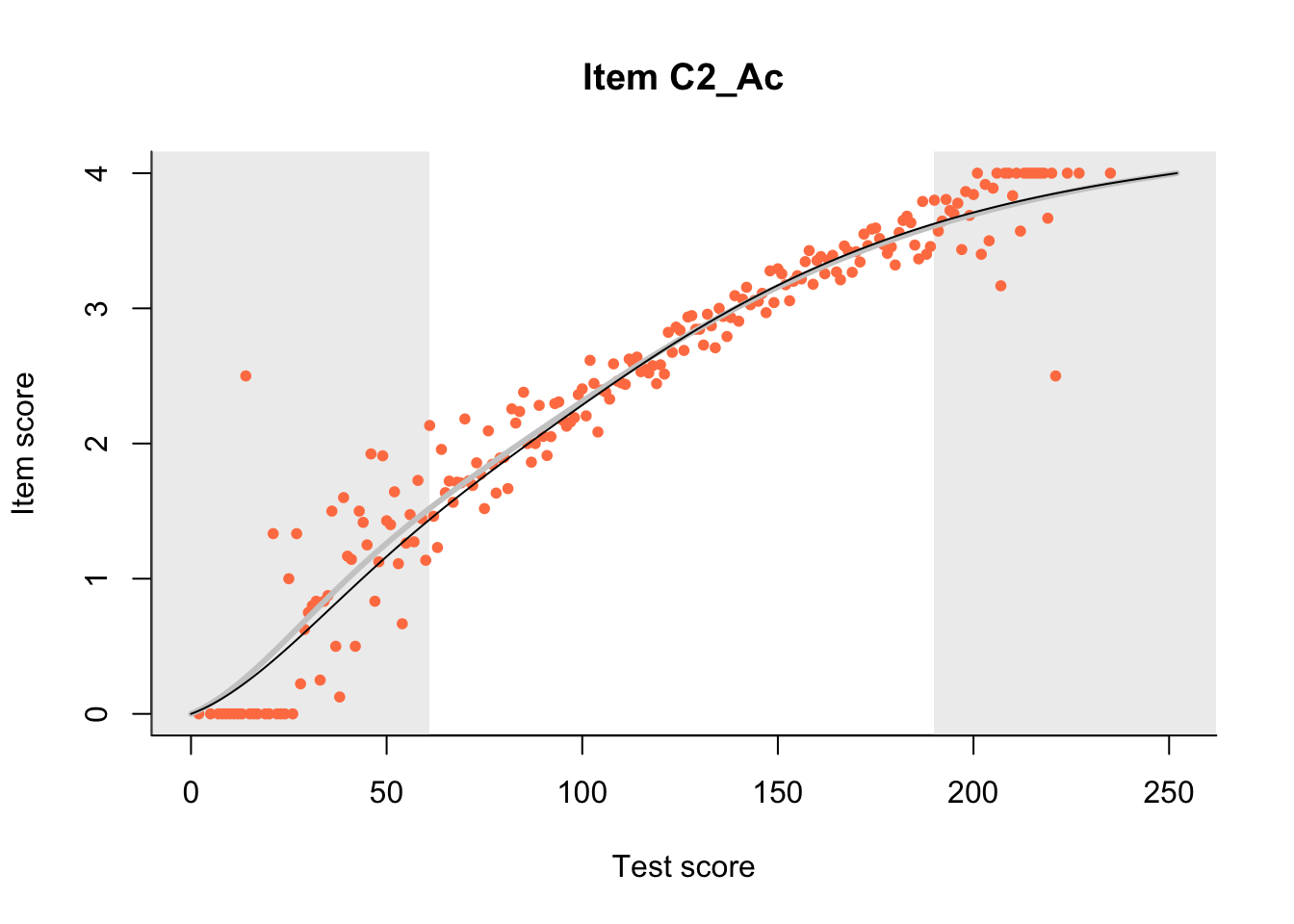
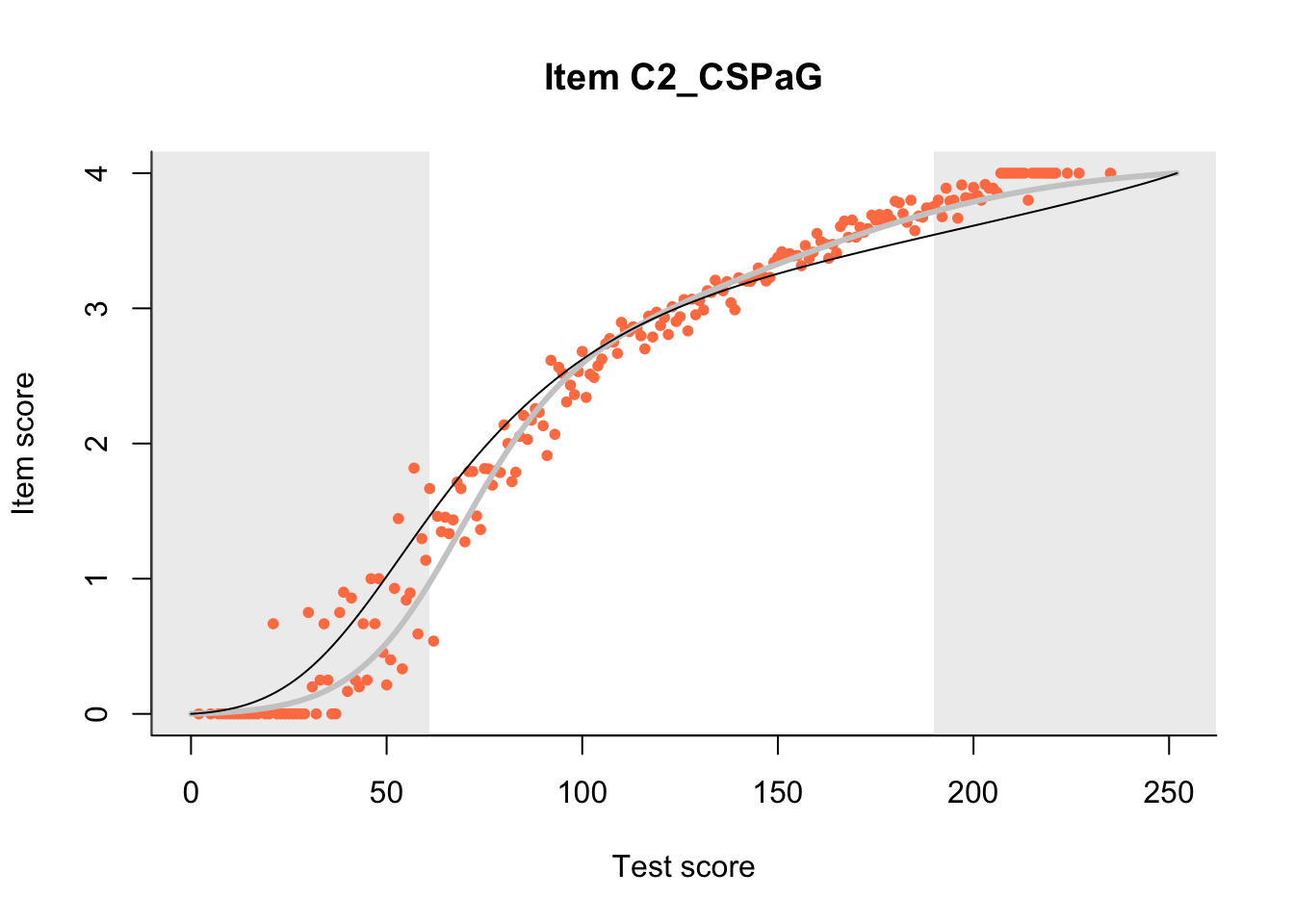
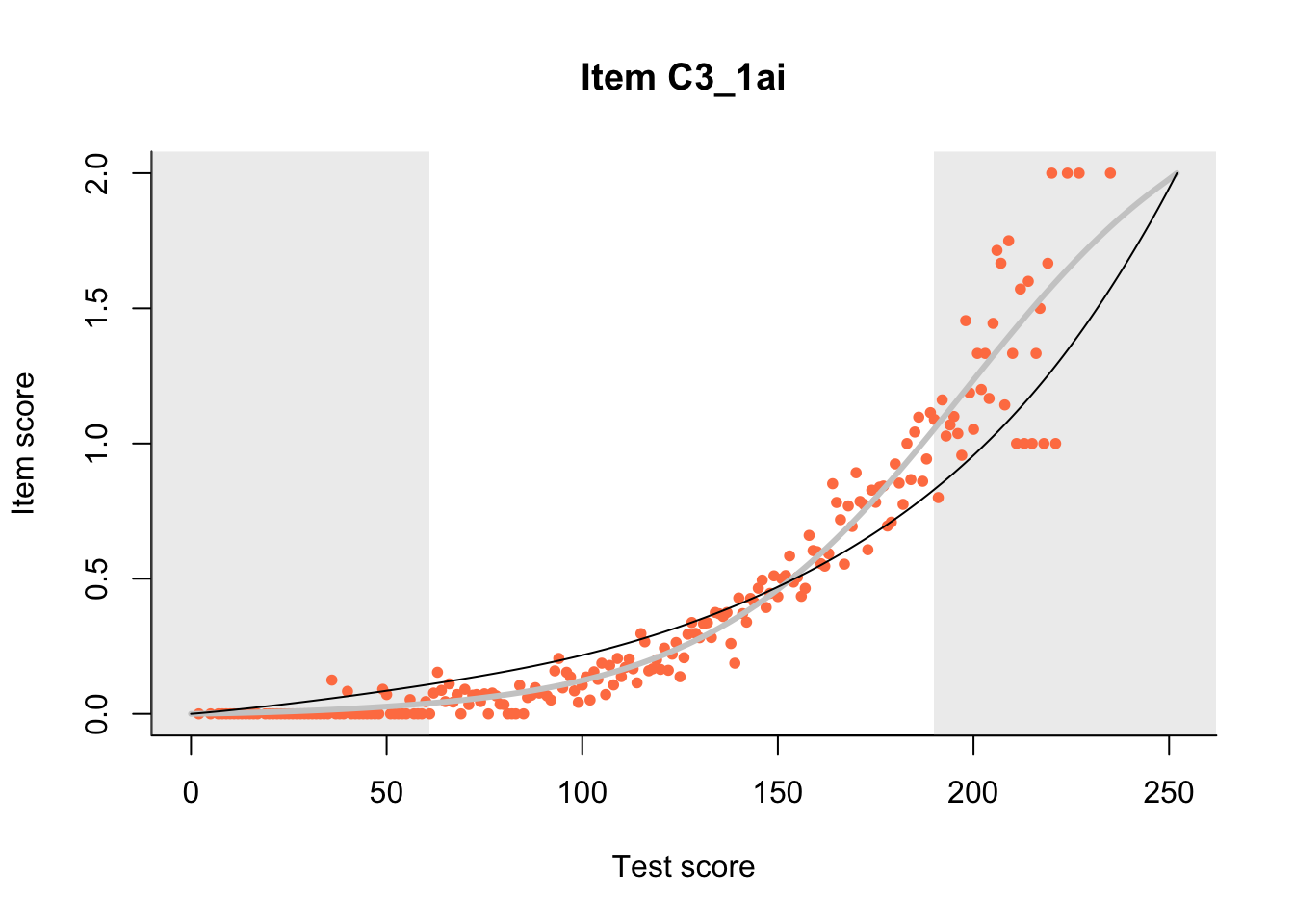
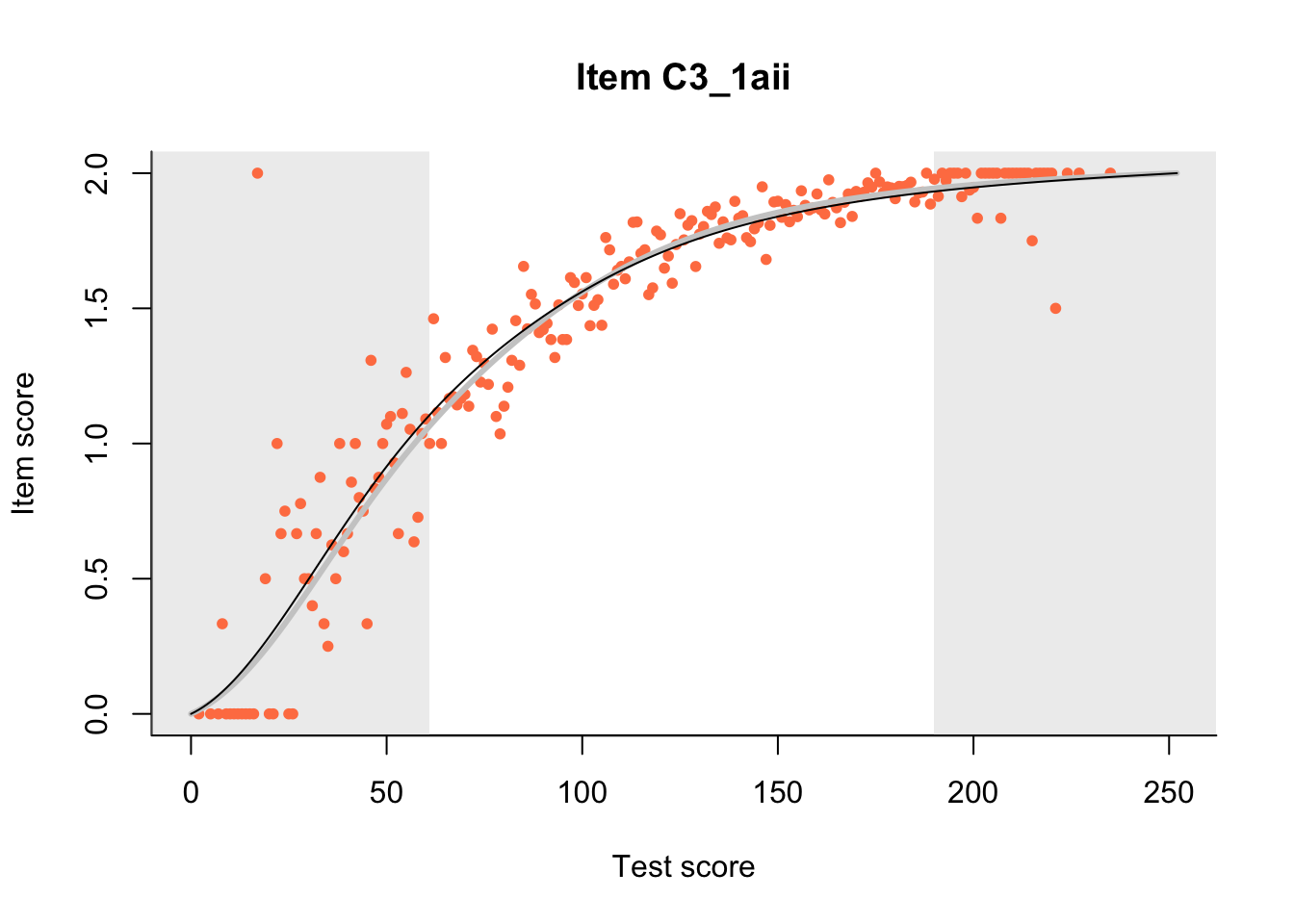
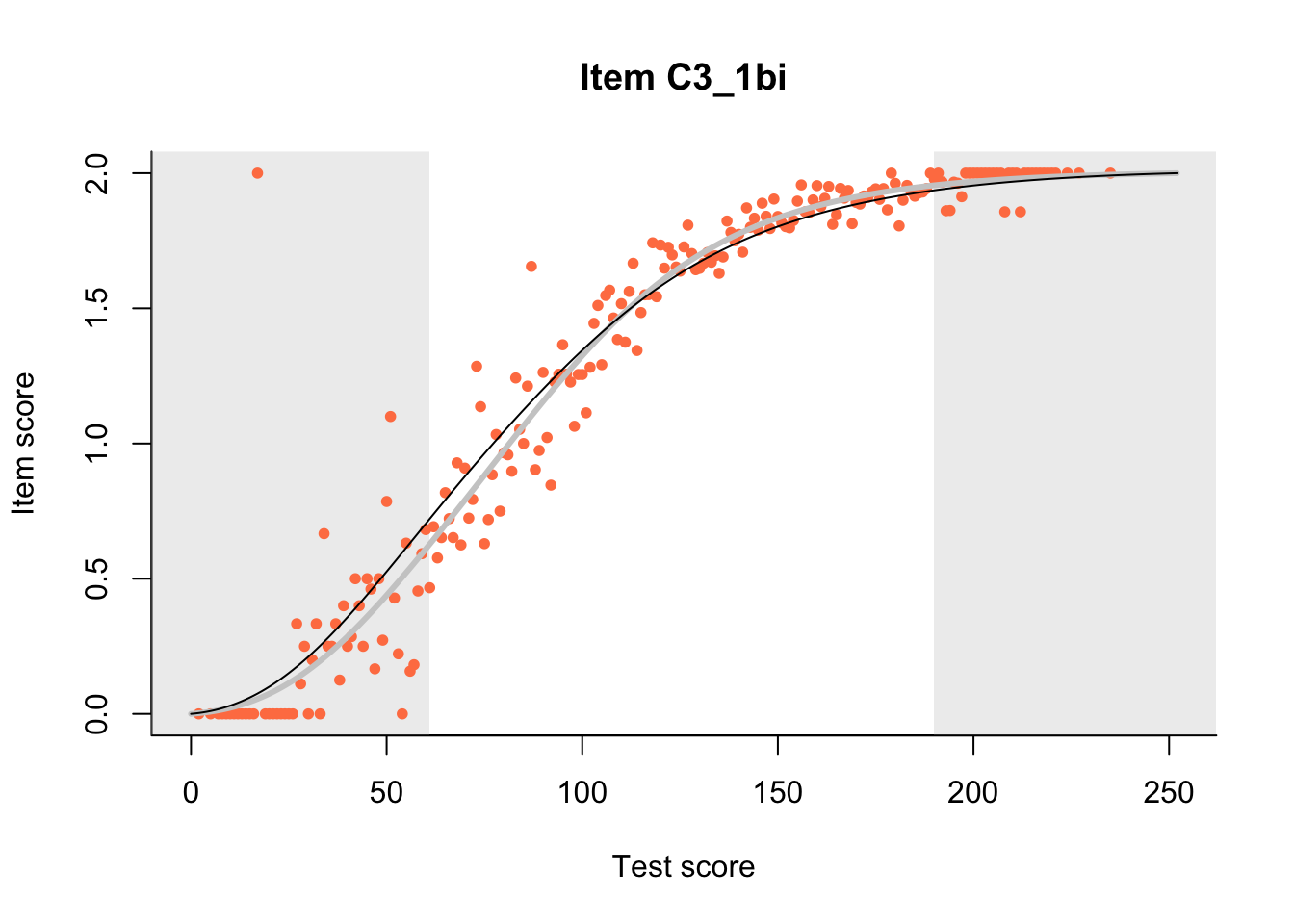
The Rasch model is shown with a thinner, darker line. The interaction model (unique to dexter) is shown with a thicker, lighter line, and the pink dots show the observed regression.
m = fit_inter(db)
n_items <- nrow(data_summary)
for(i in 1:n_items){
plot(m, data_summary$Variable[i], show.observed=TRUE)
}
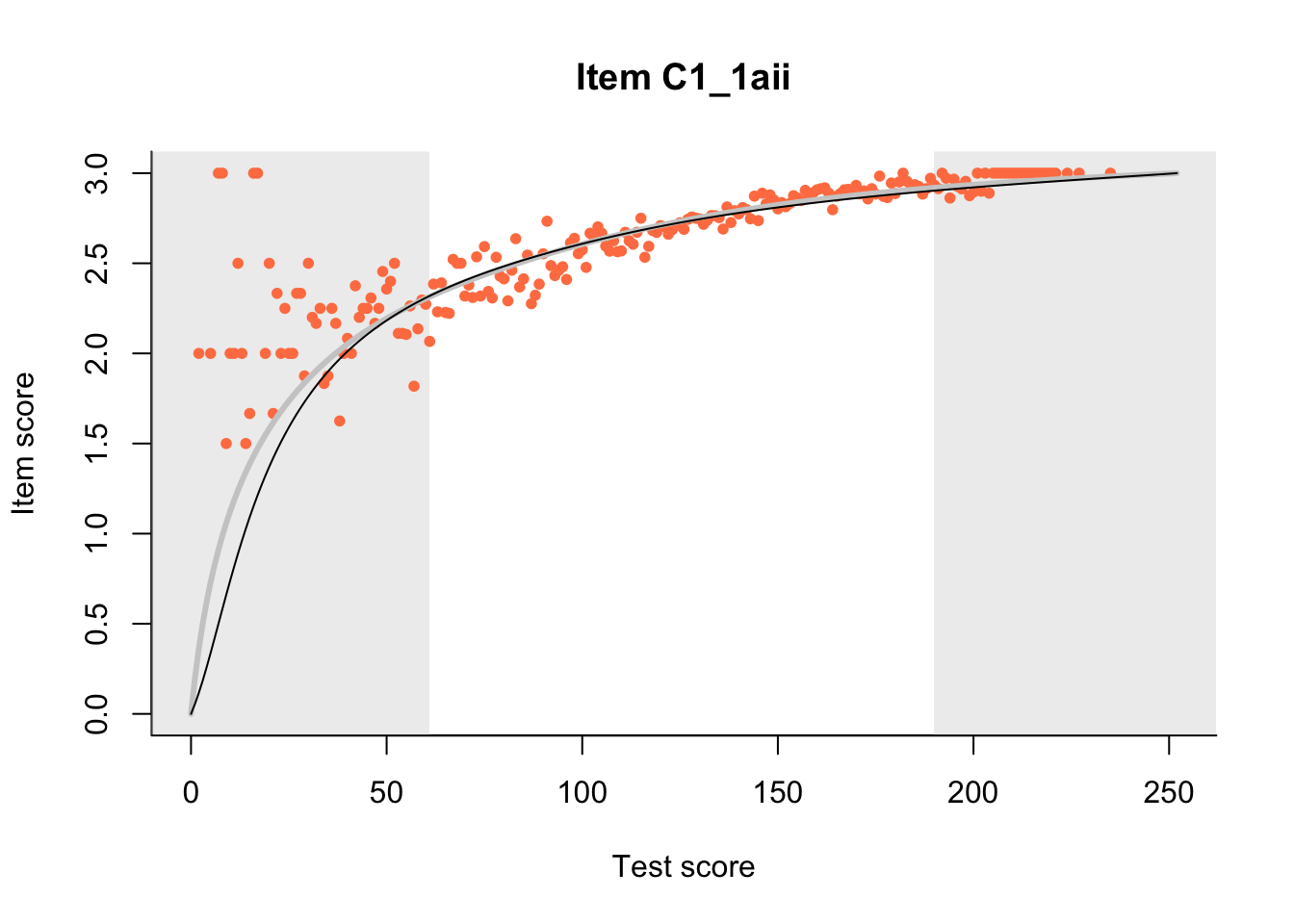
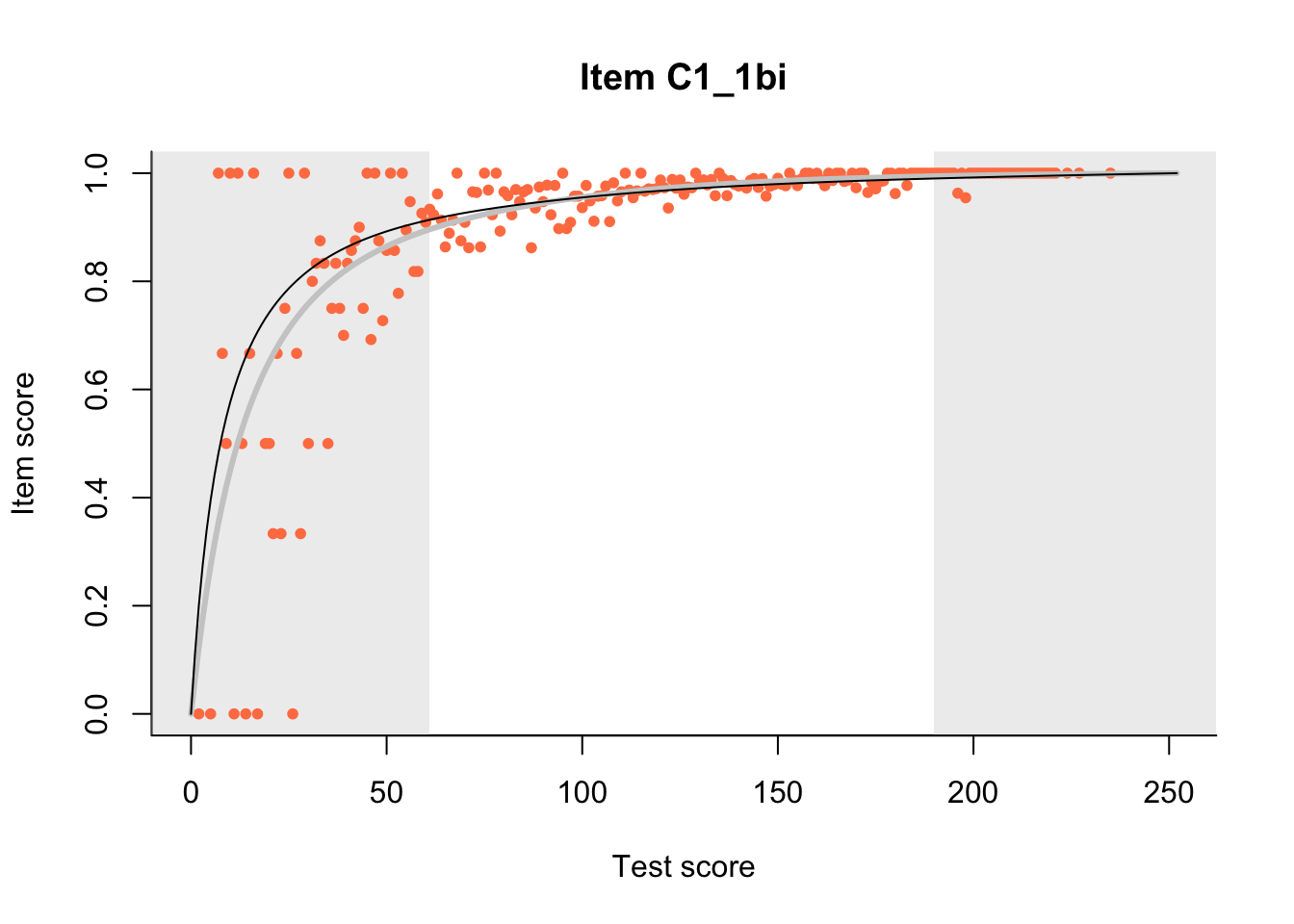
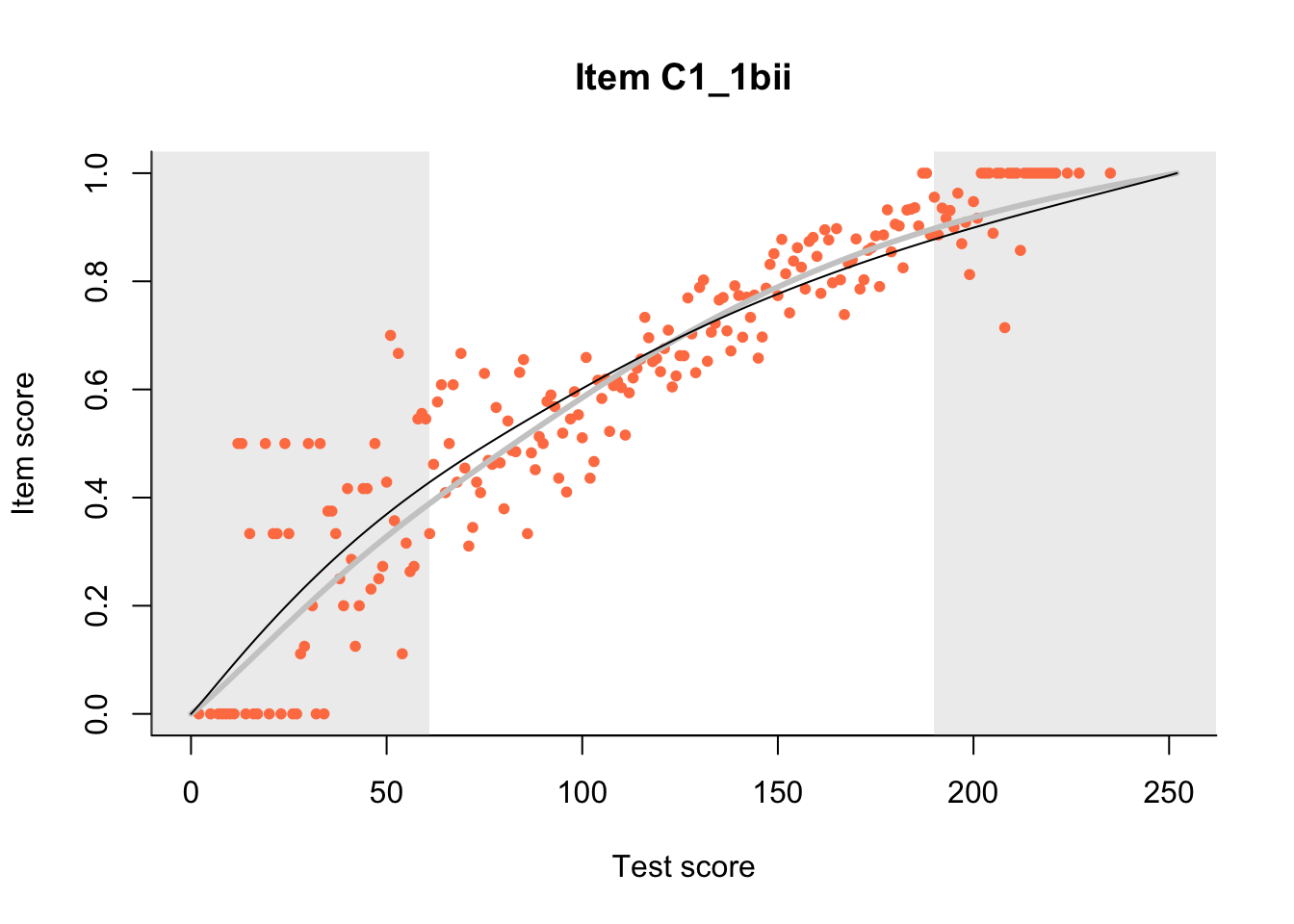
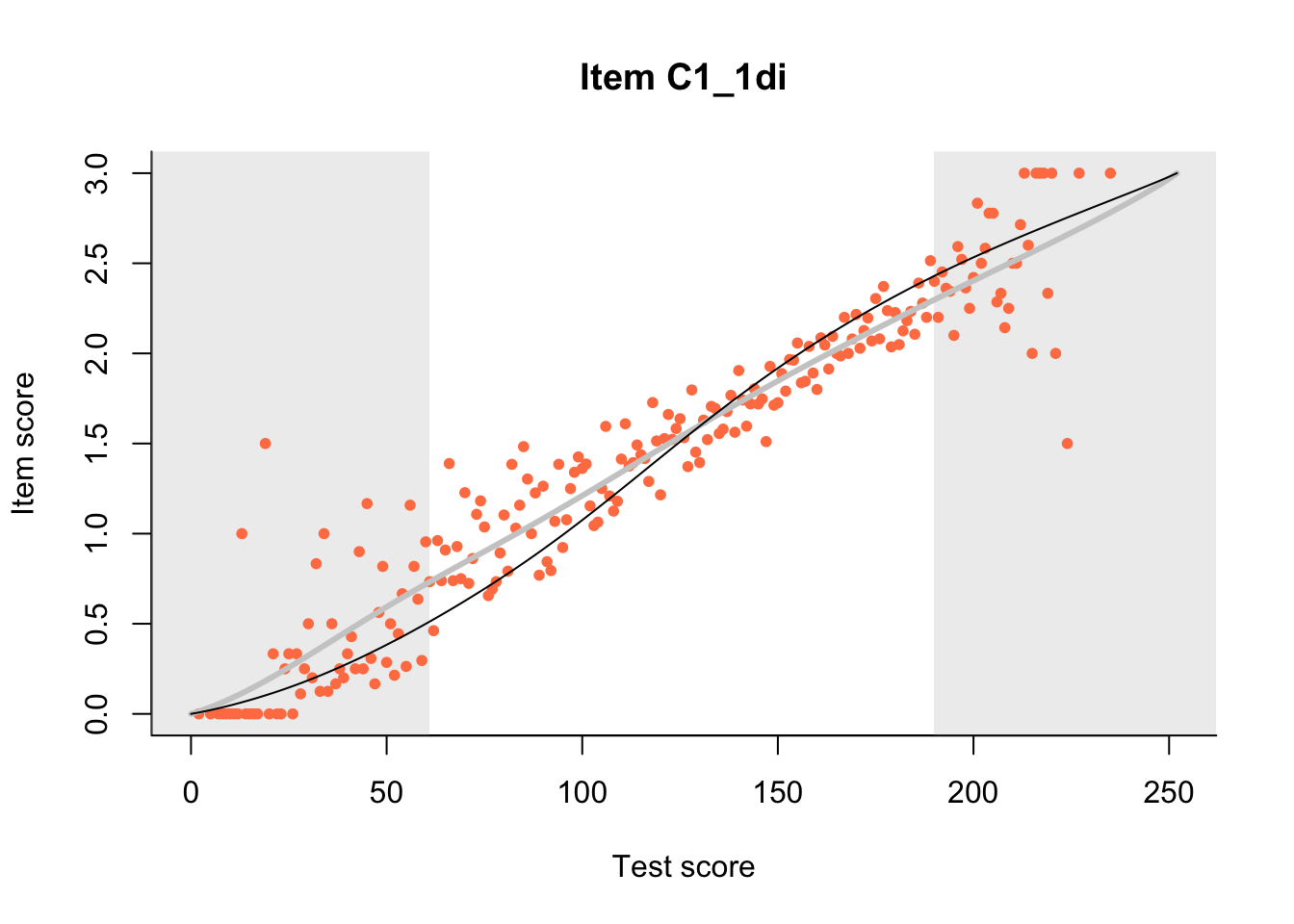
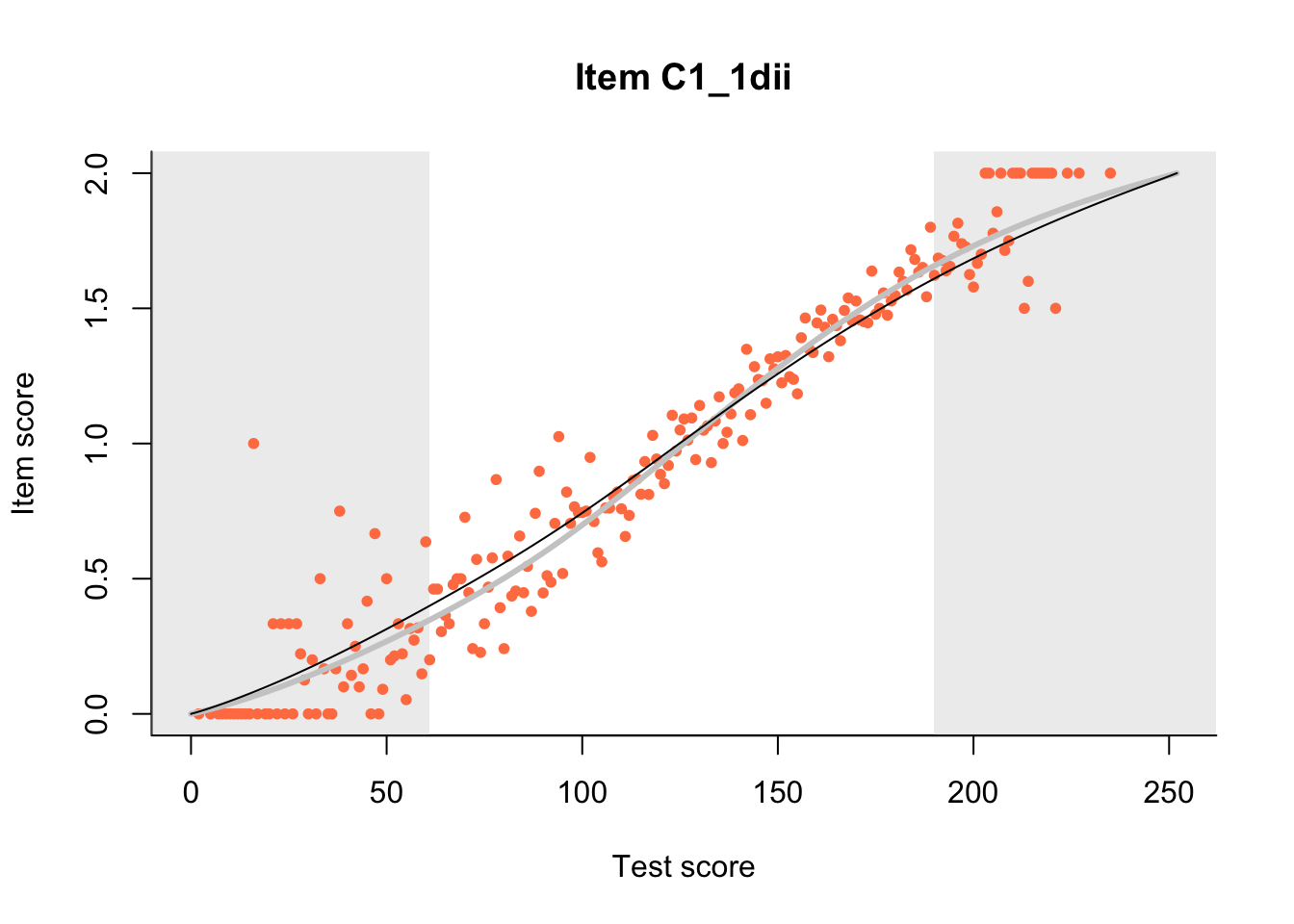
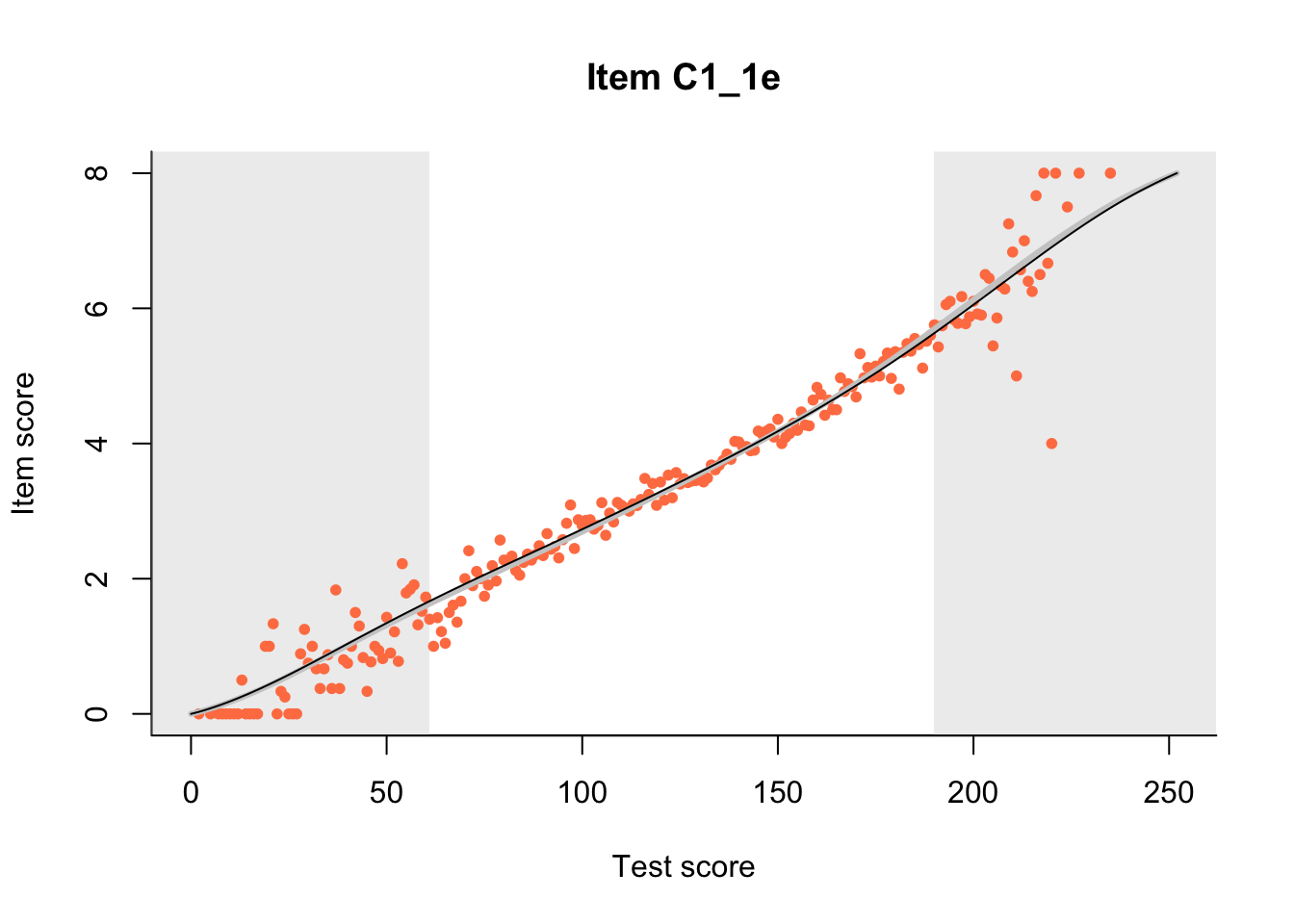
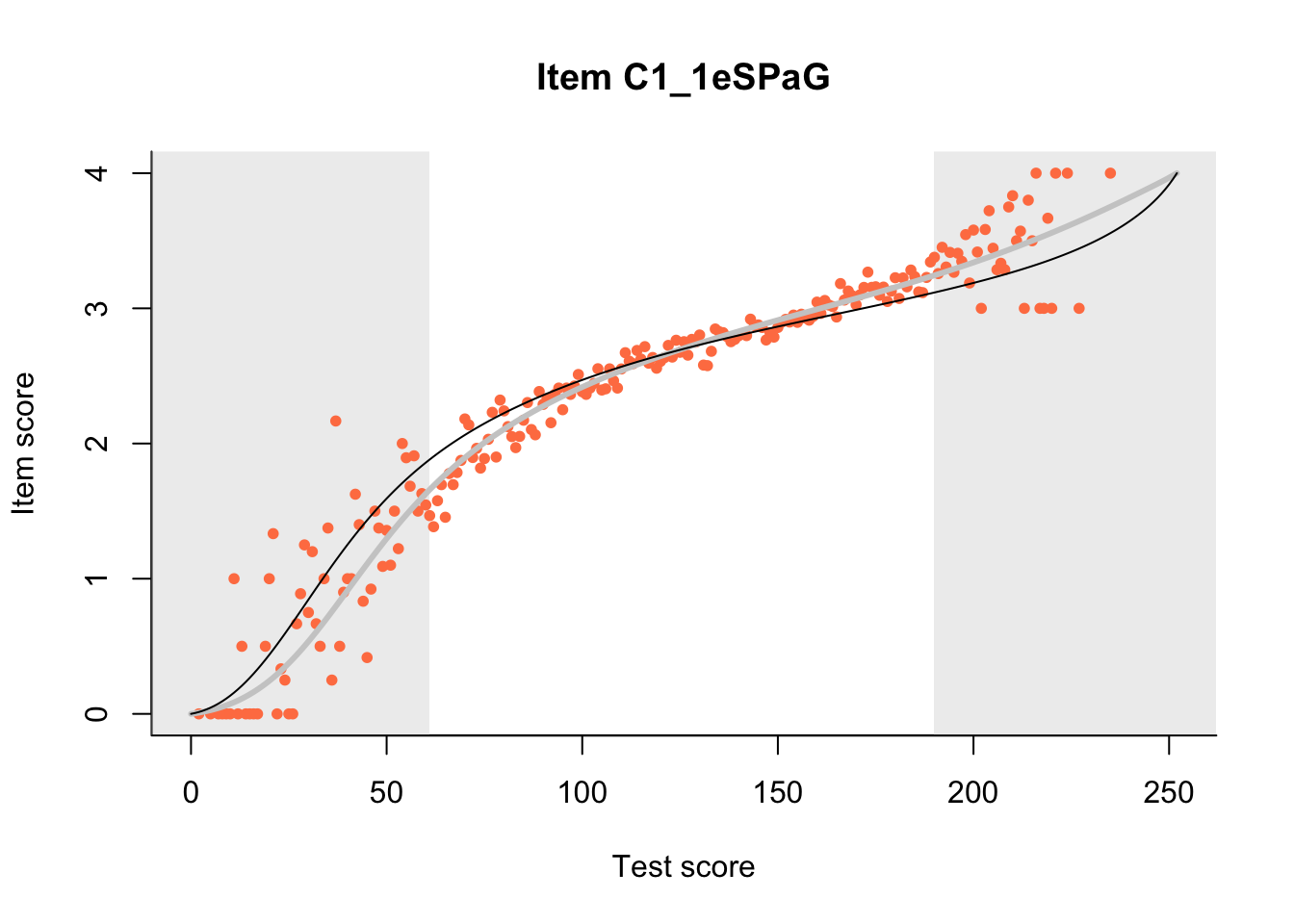
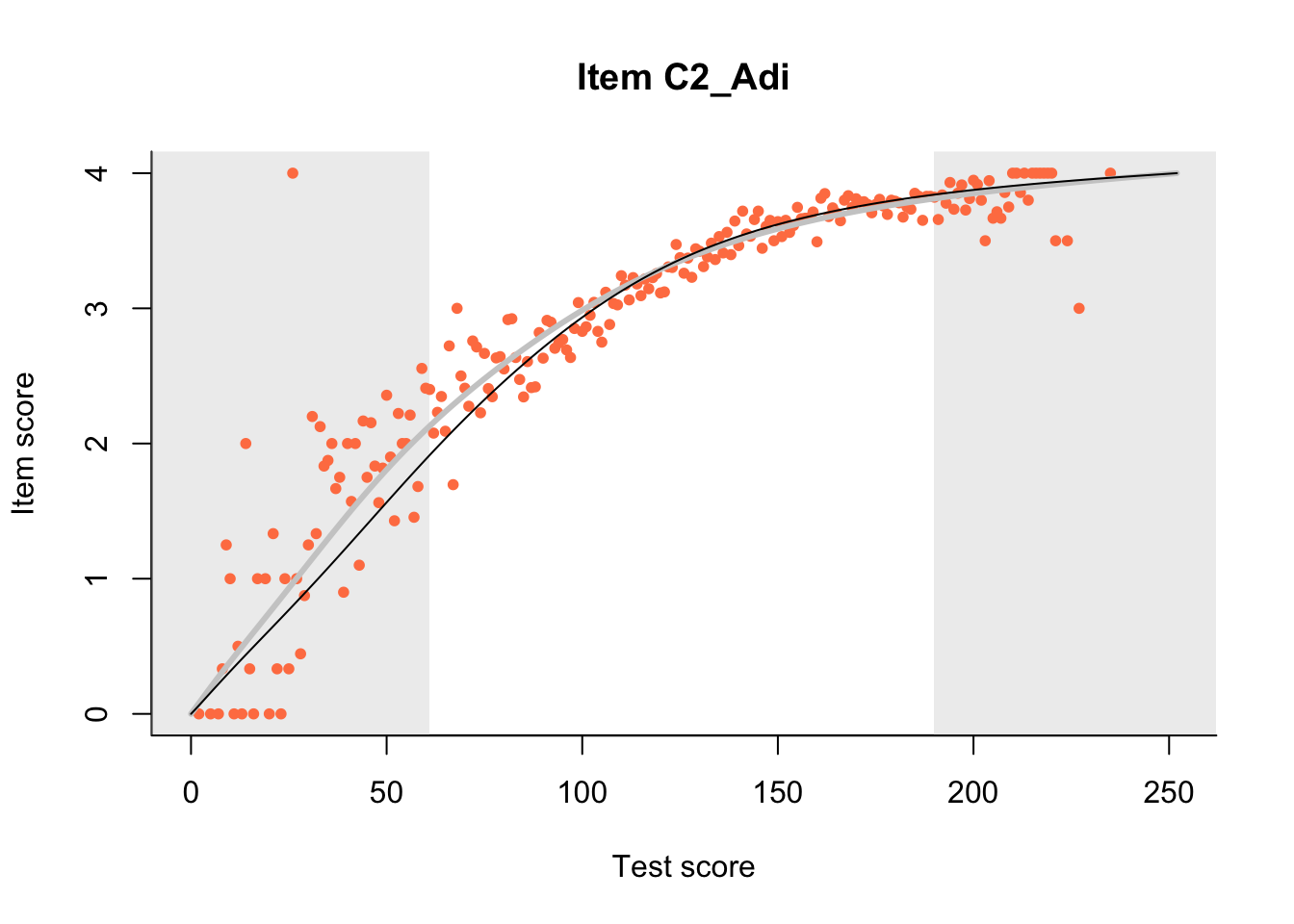
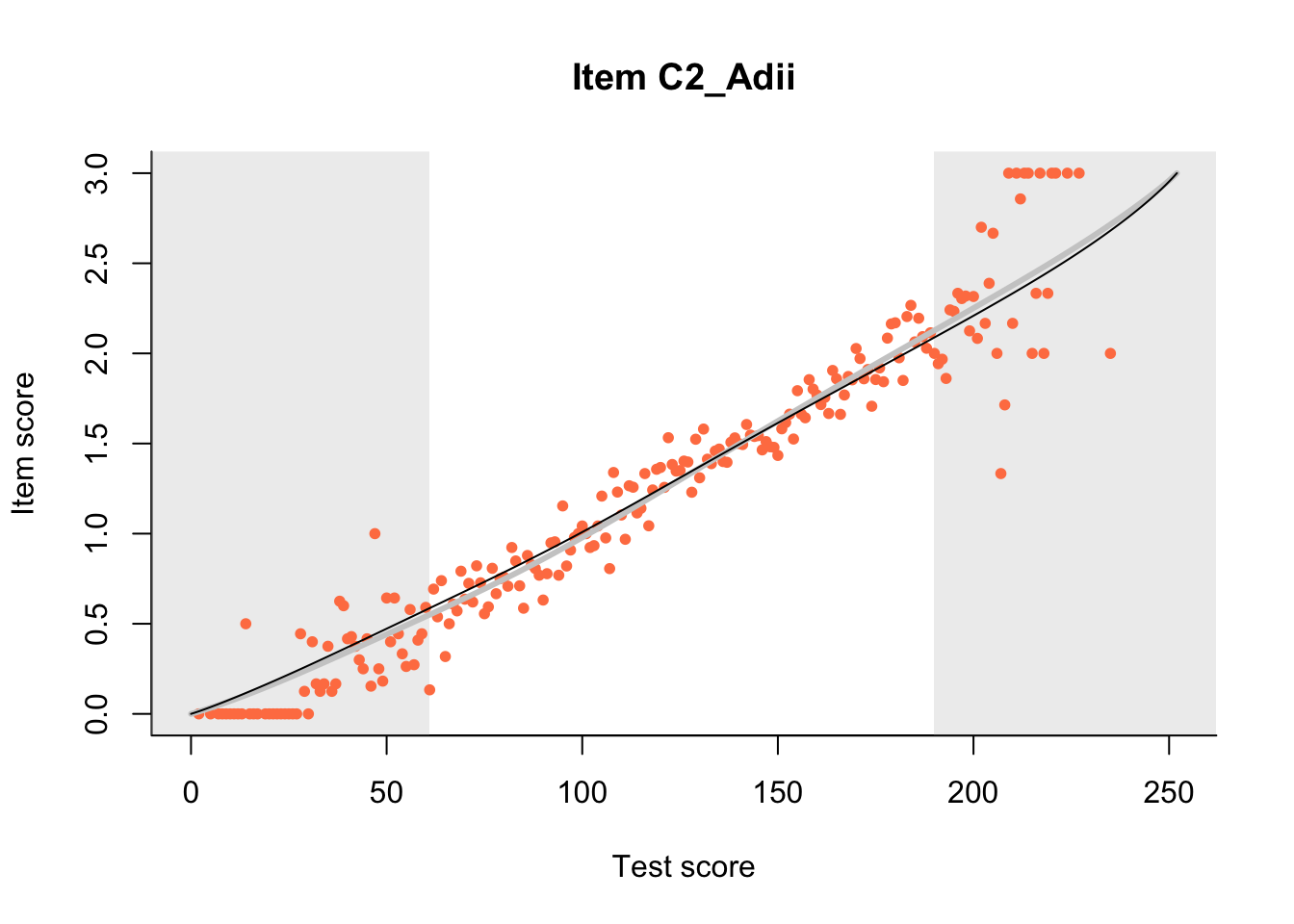
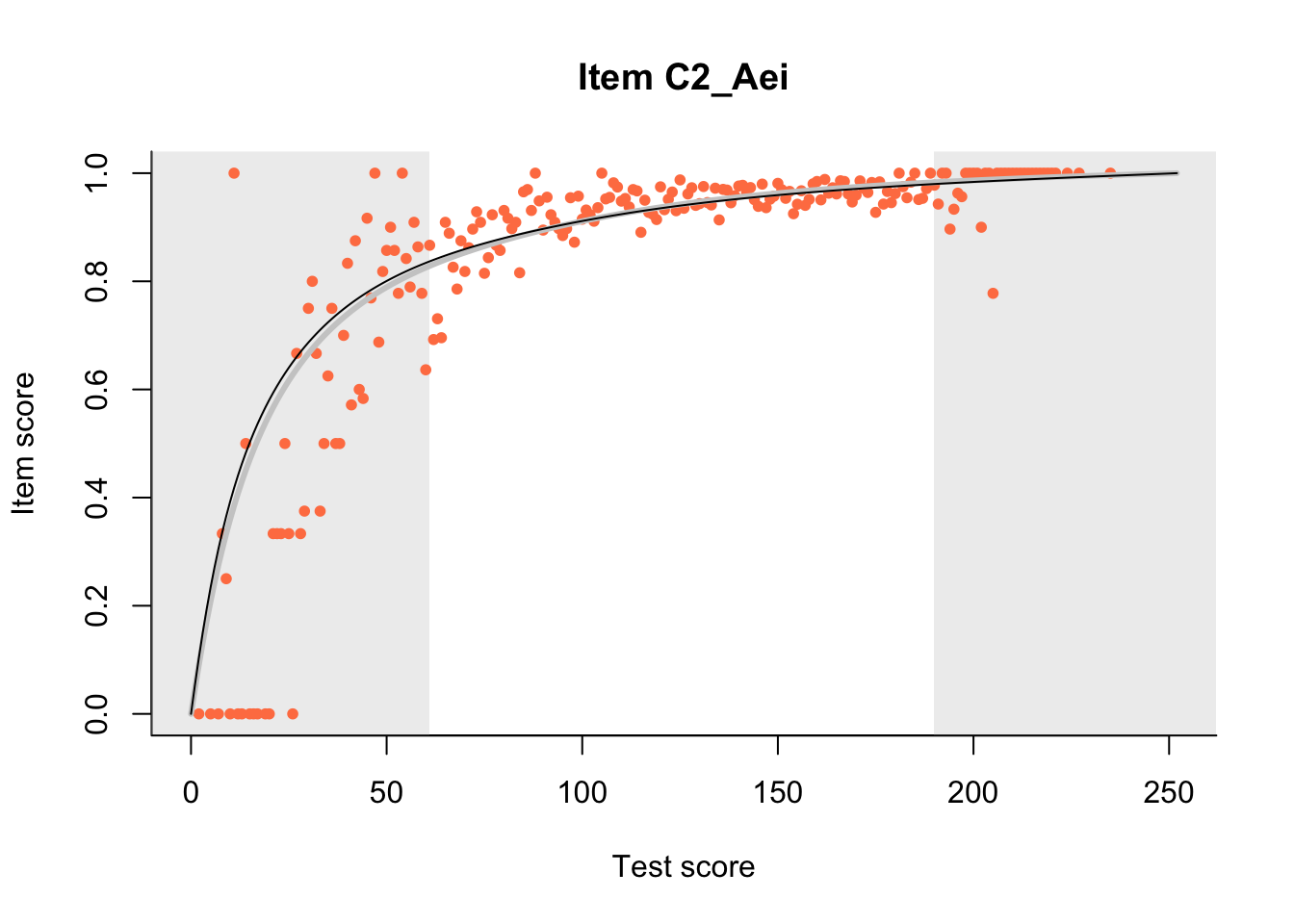
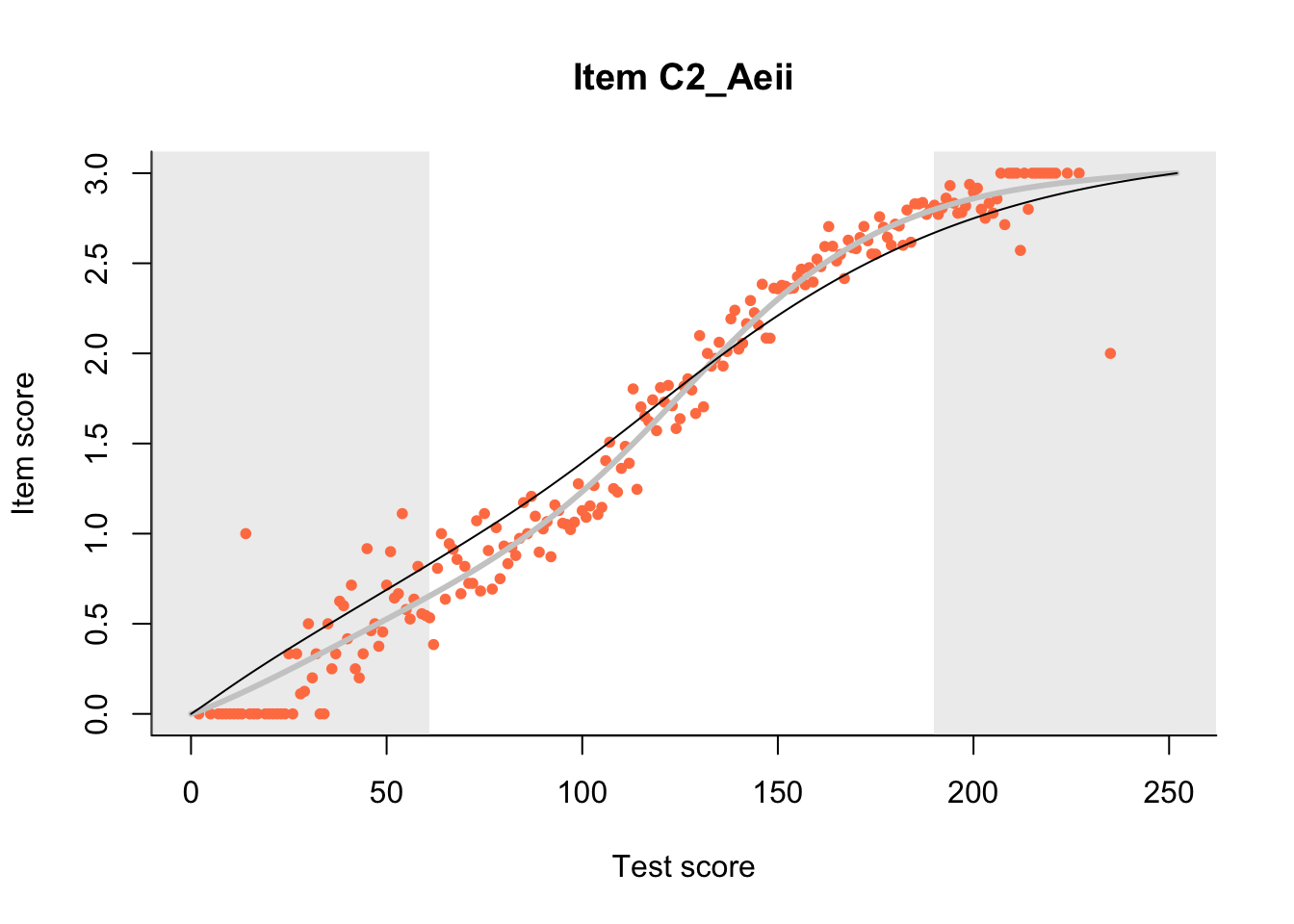
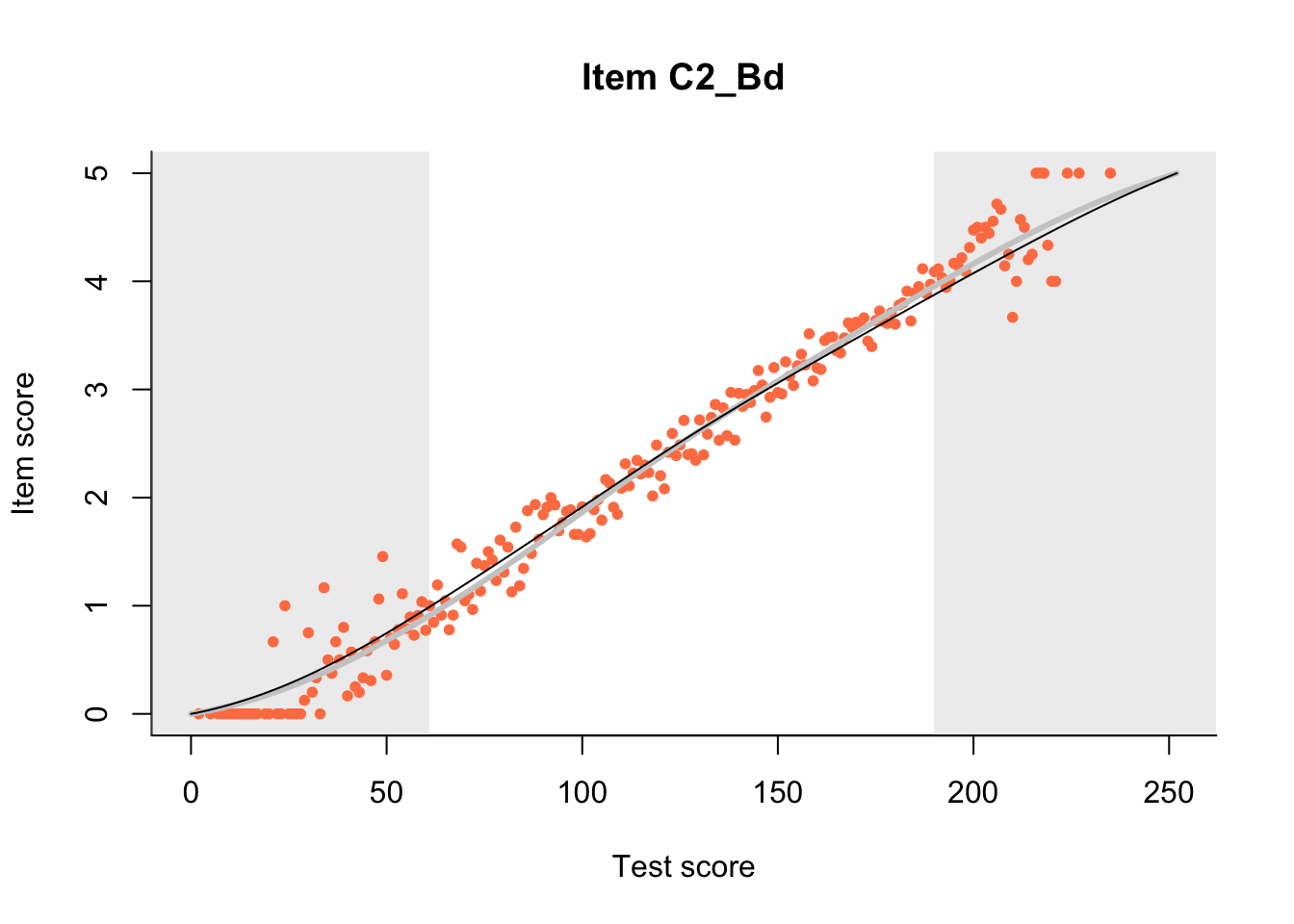
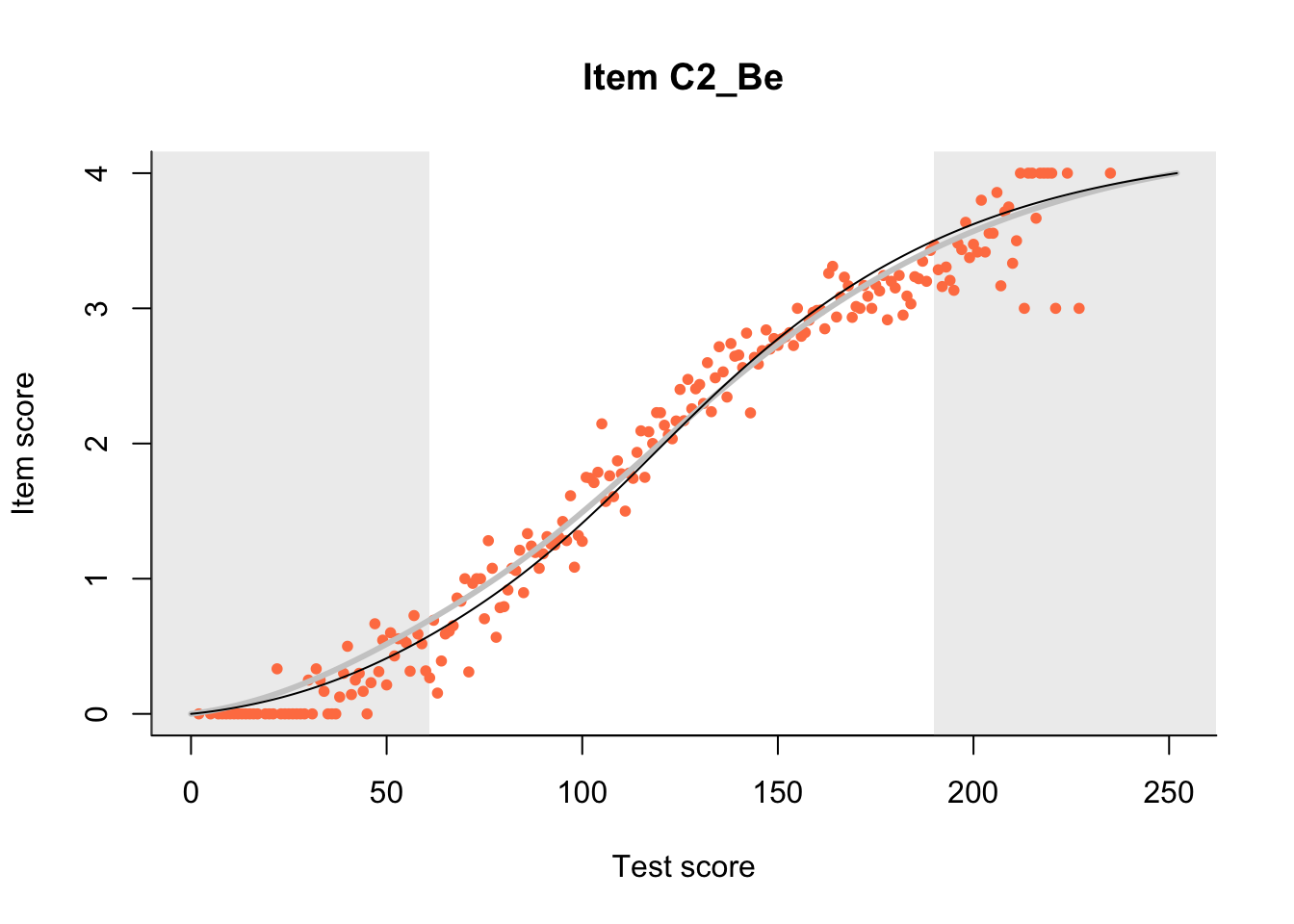
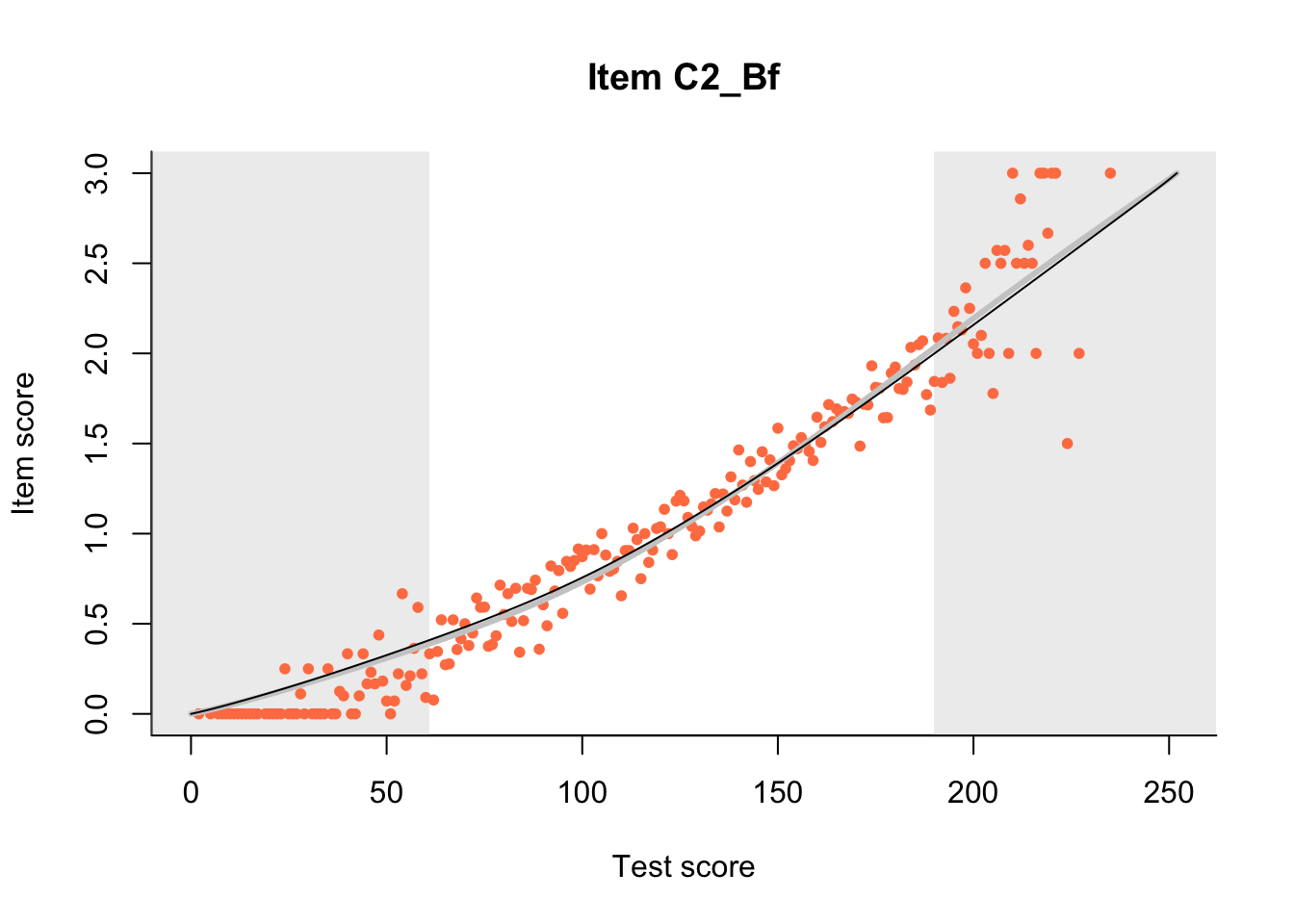
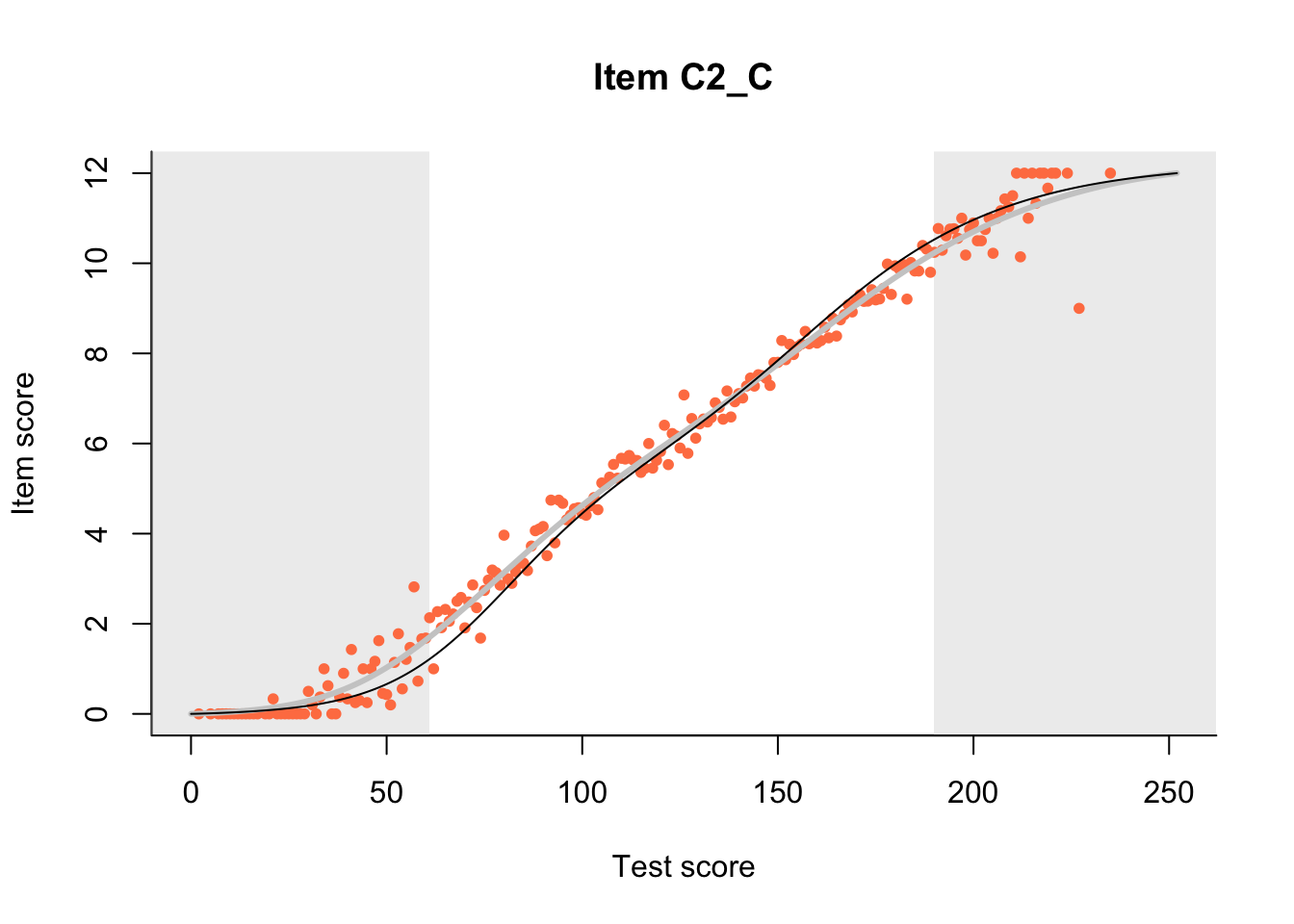
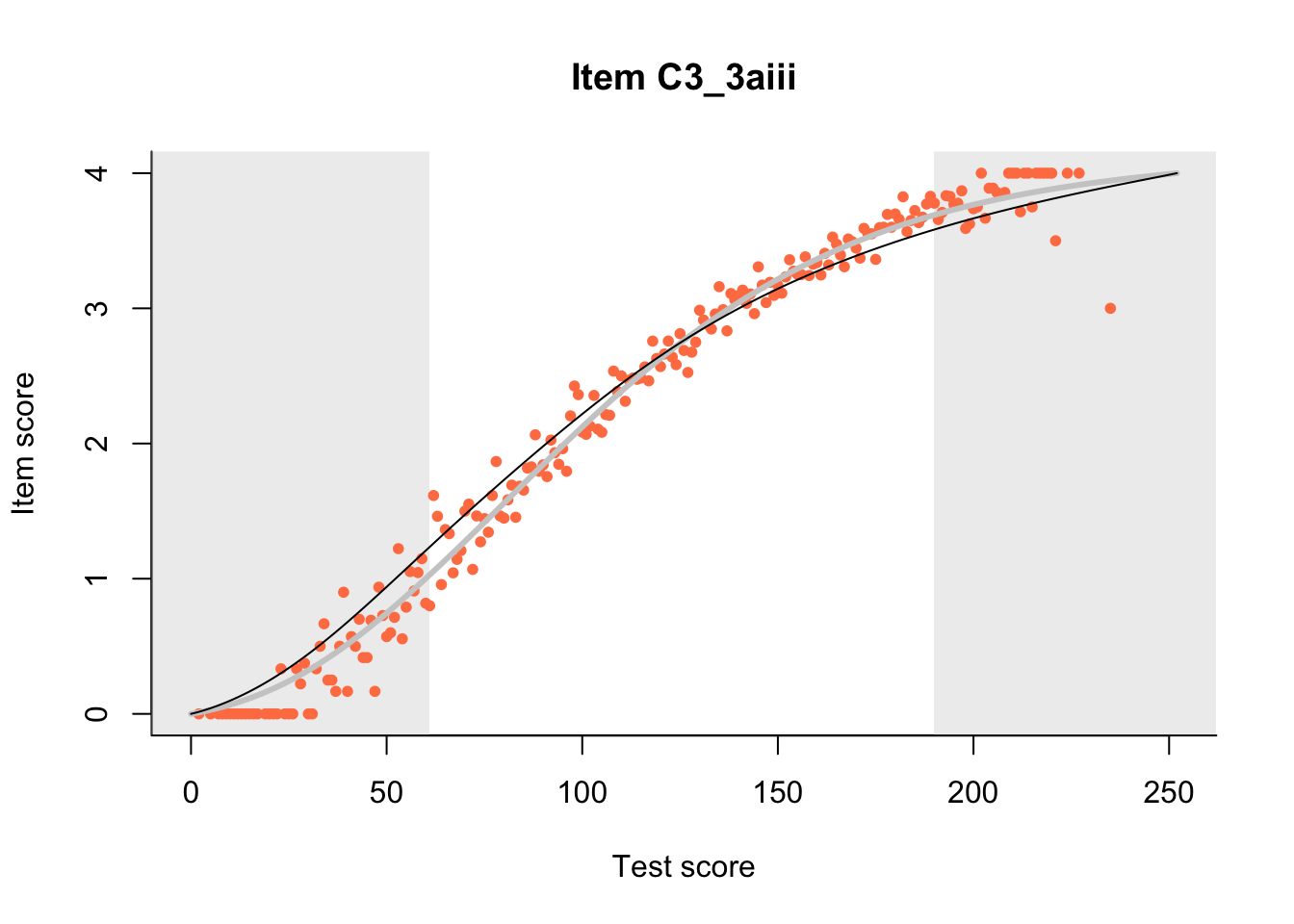
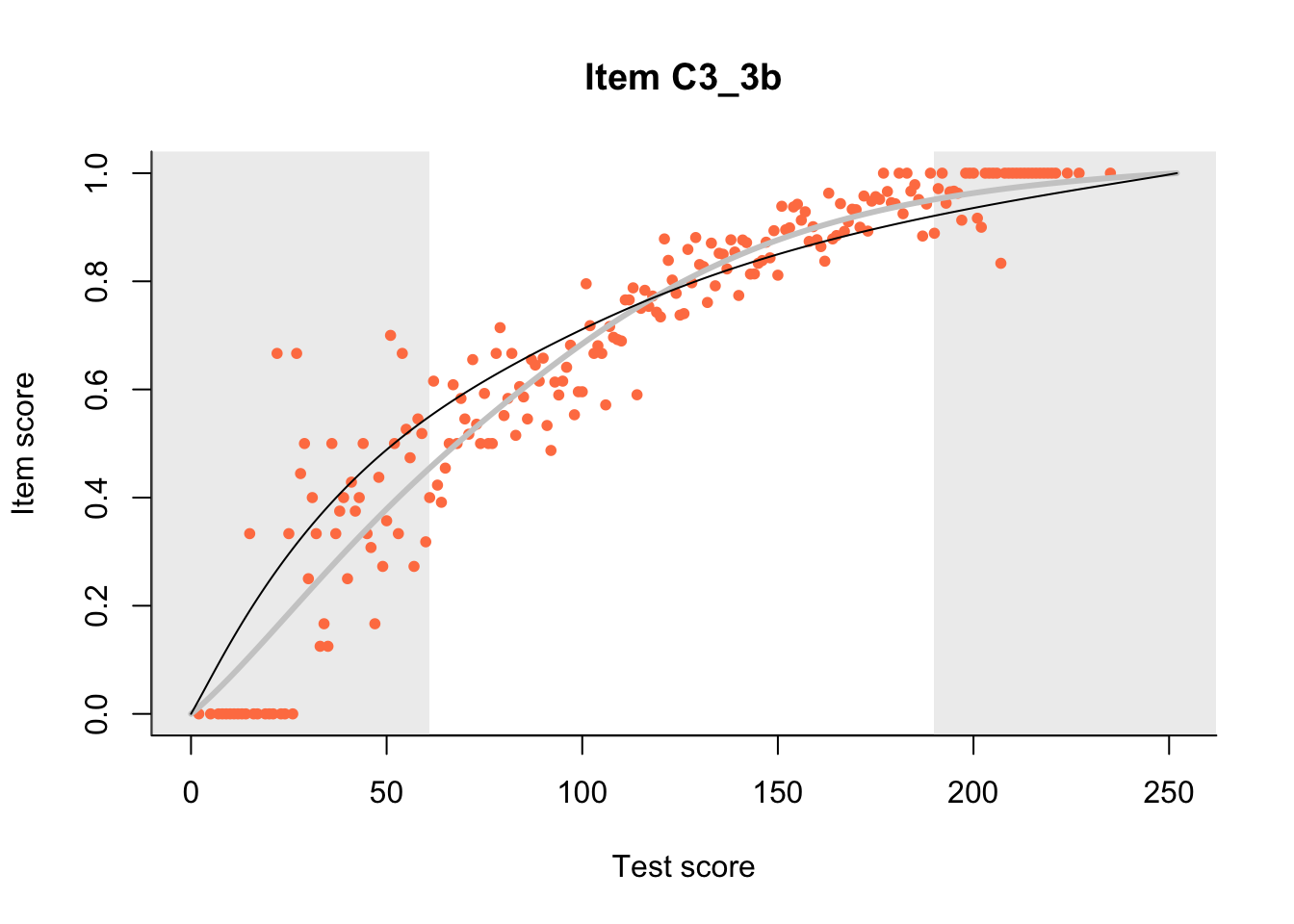
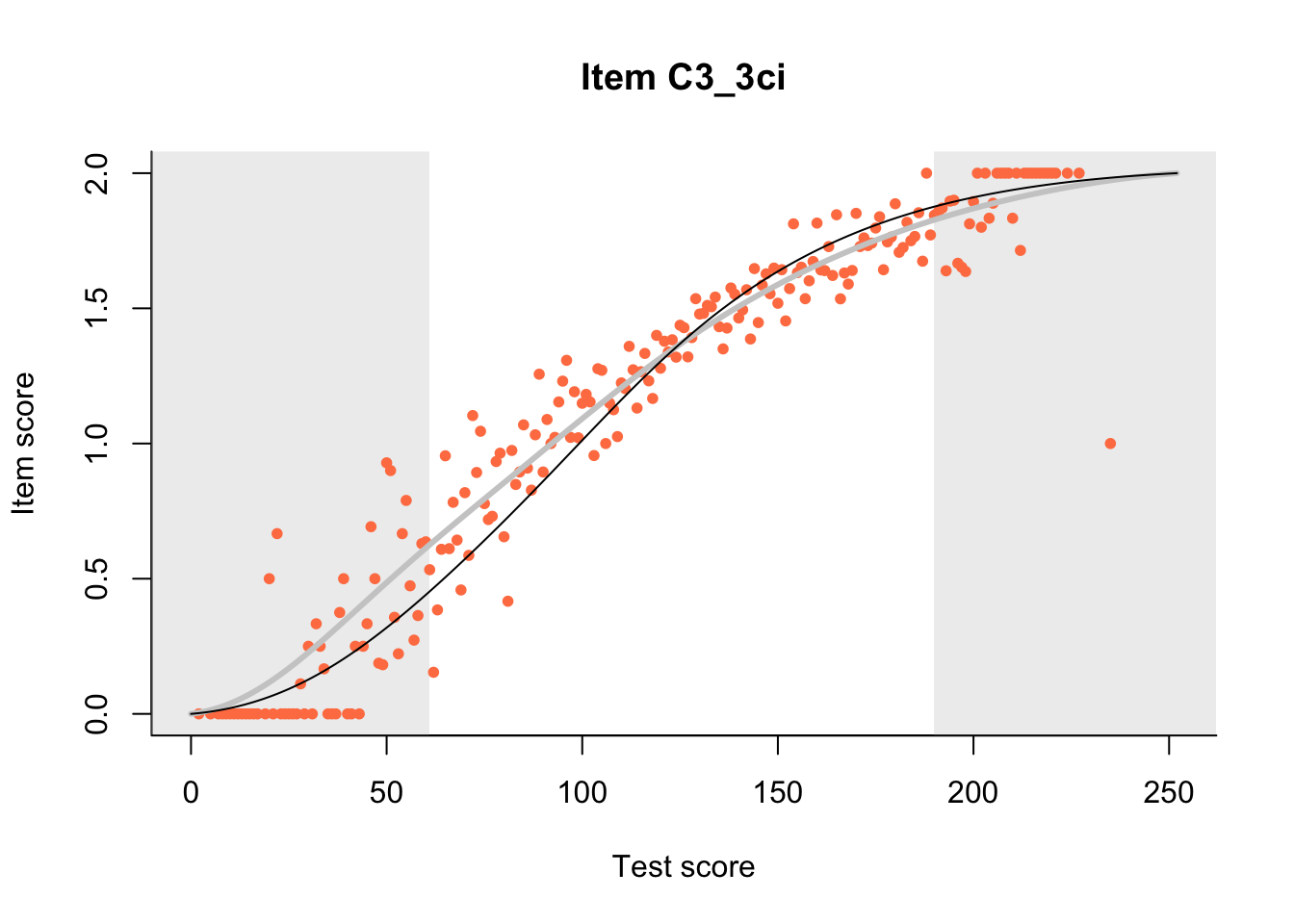
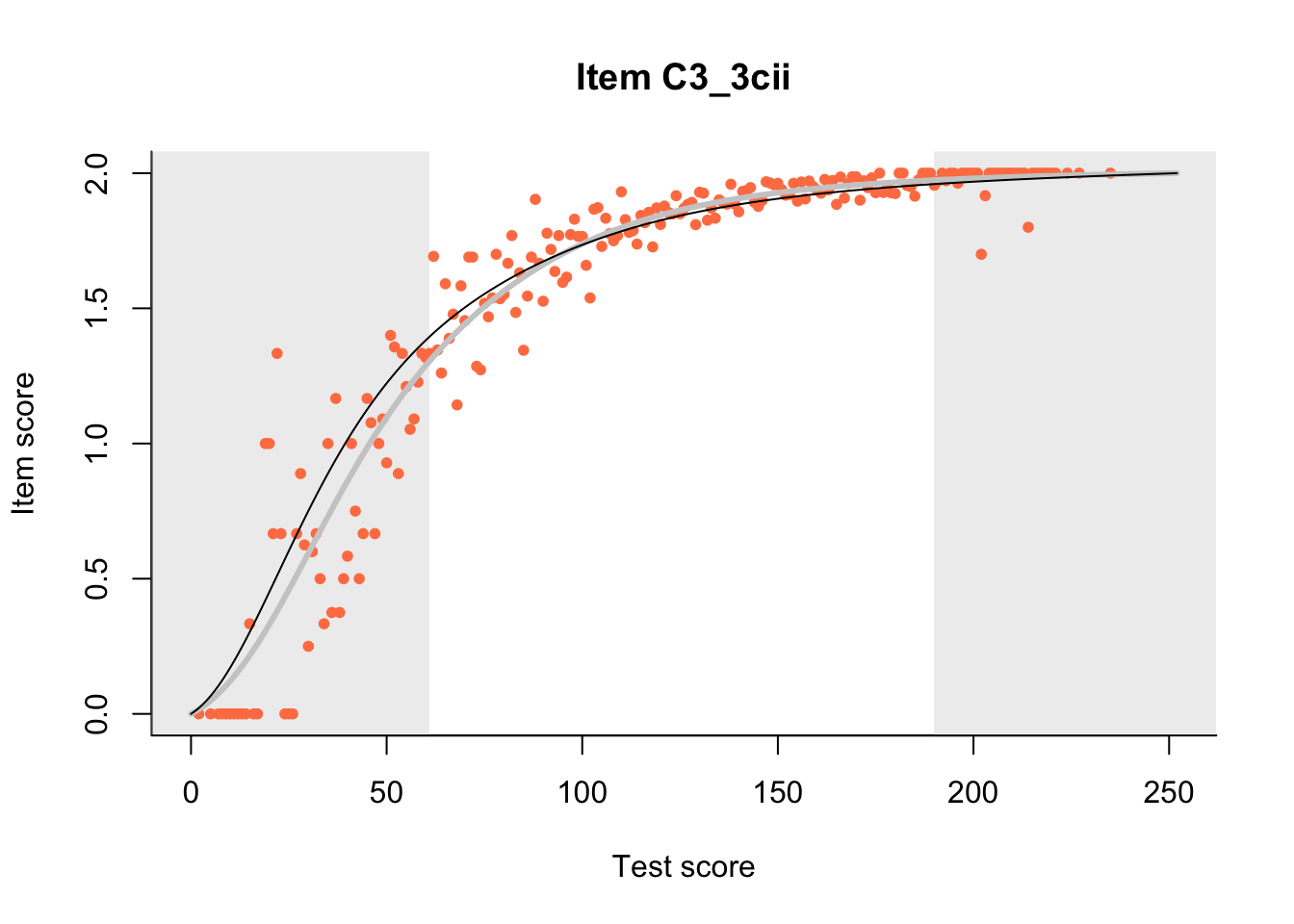
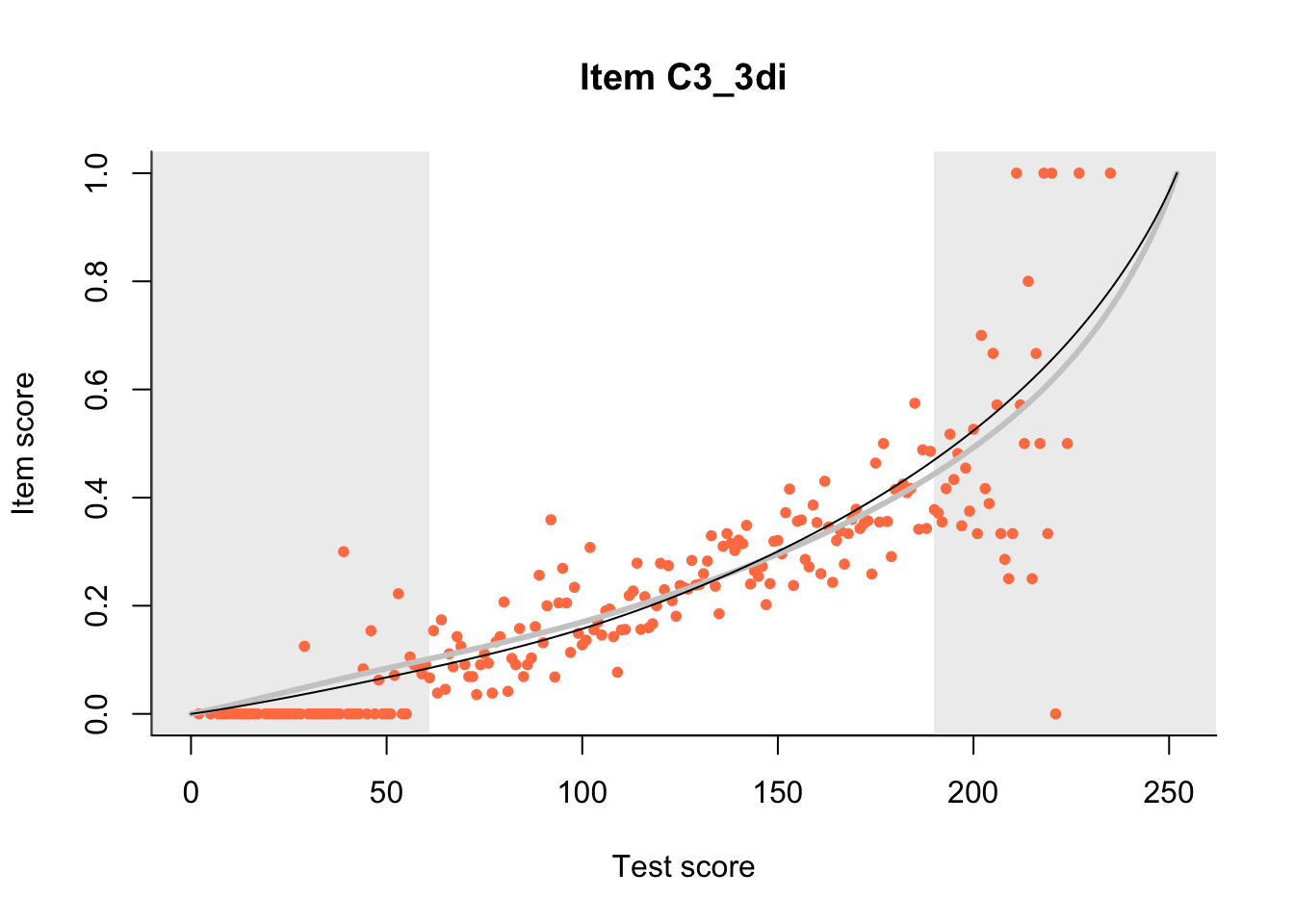
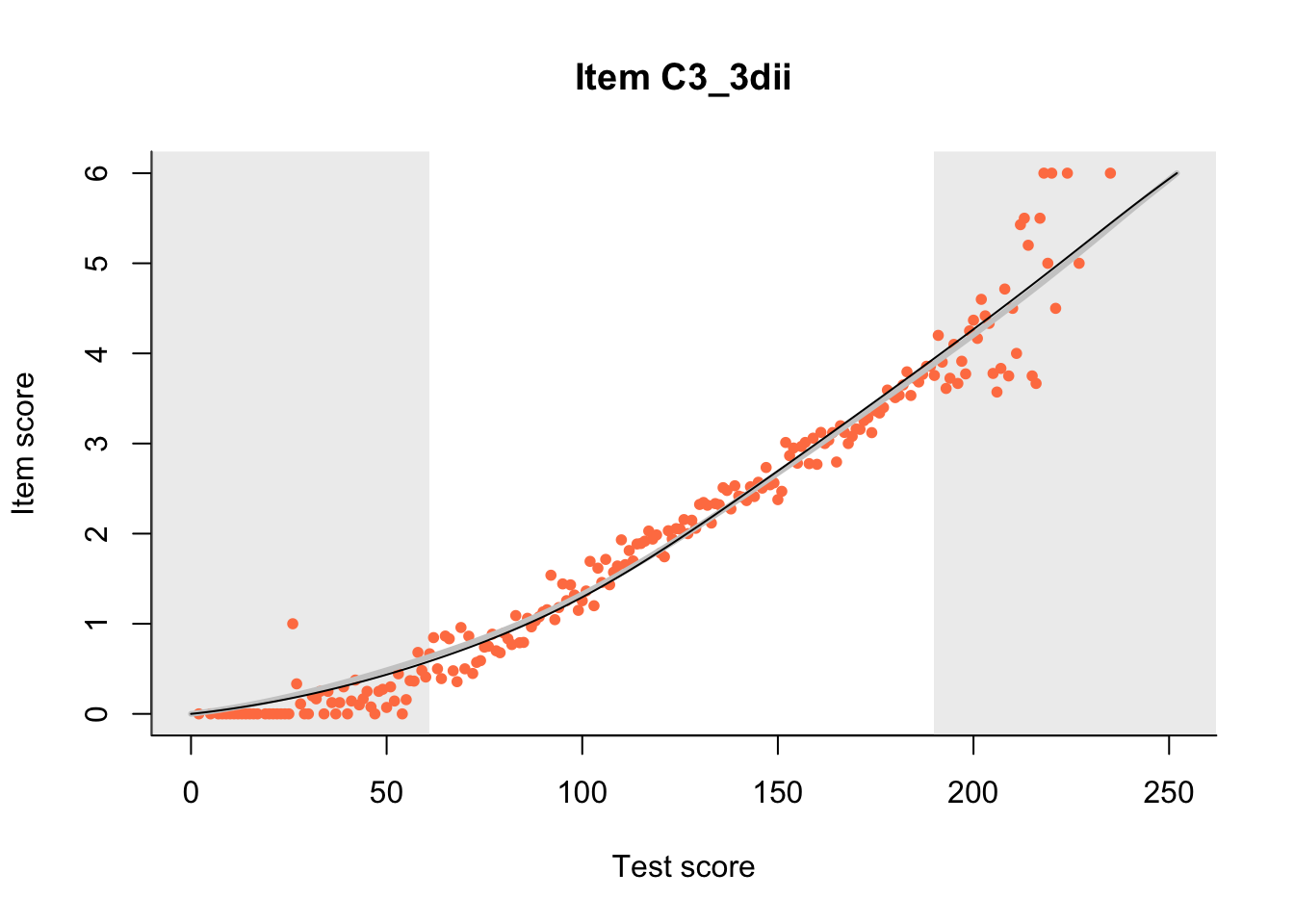
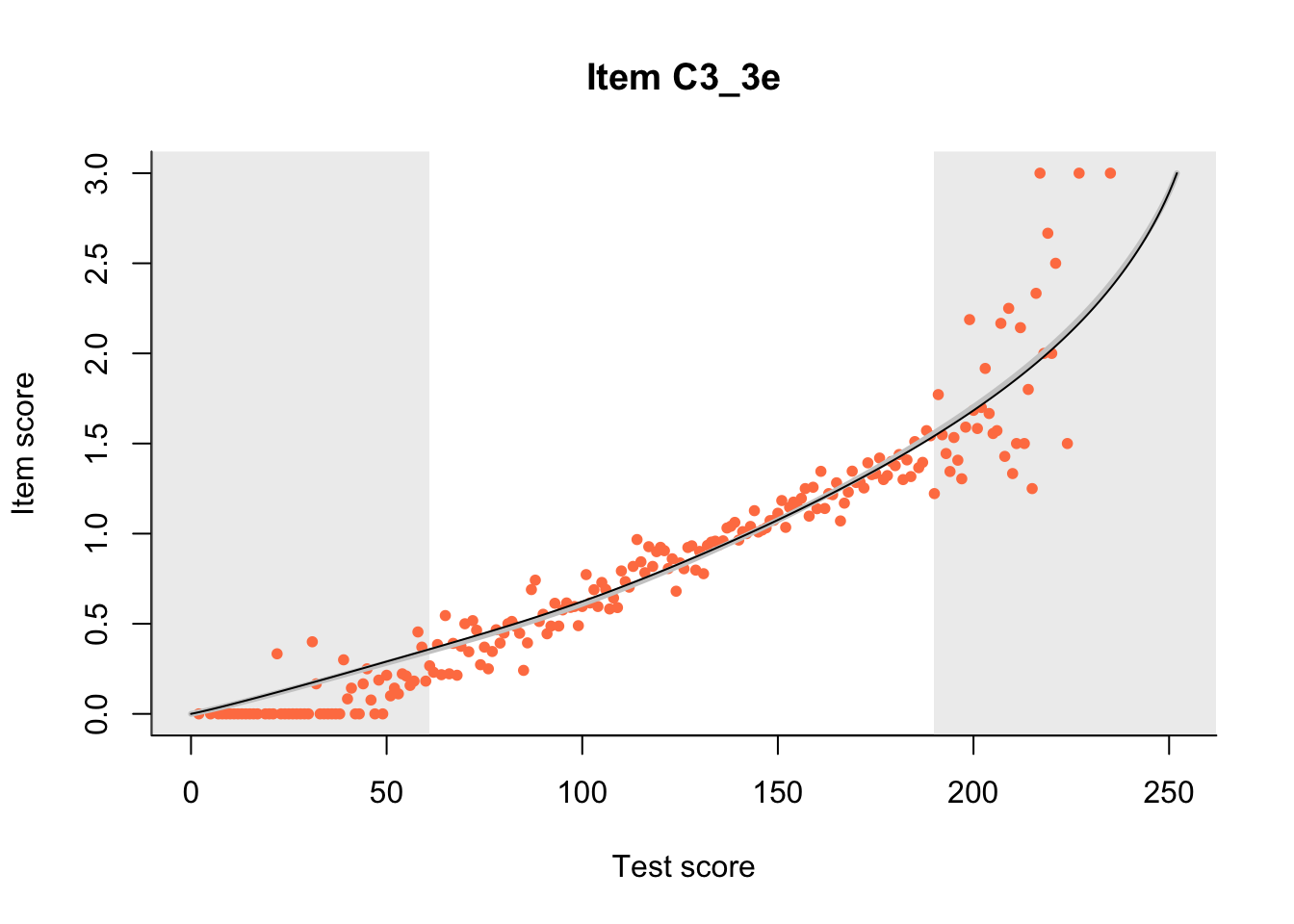
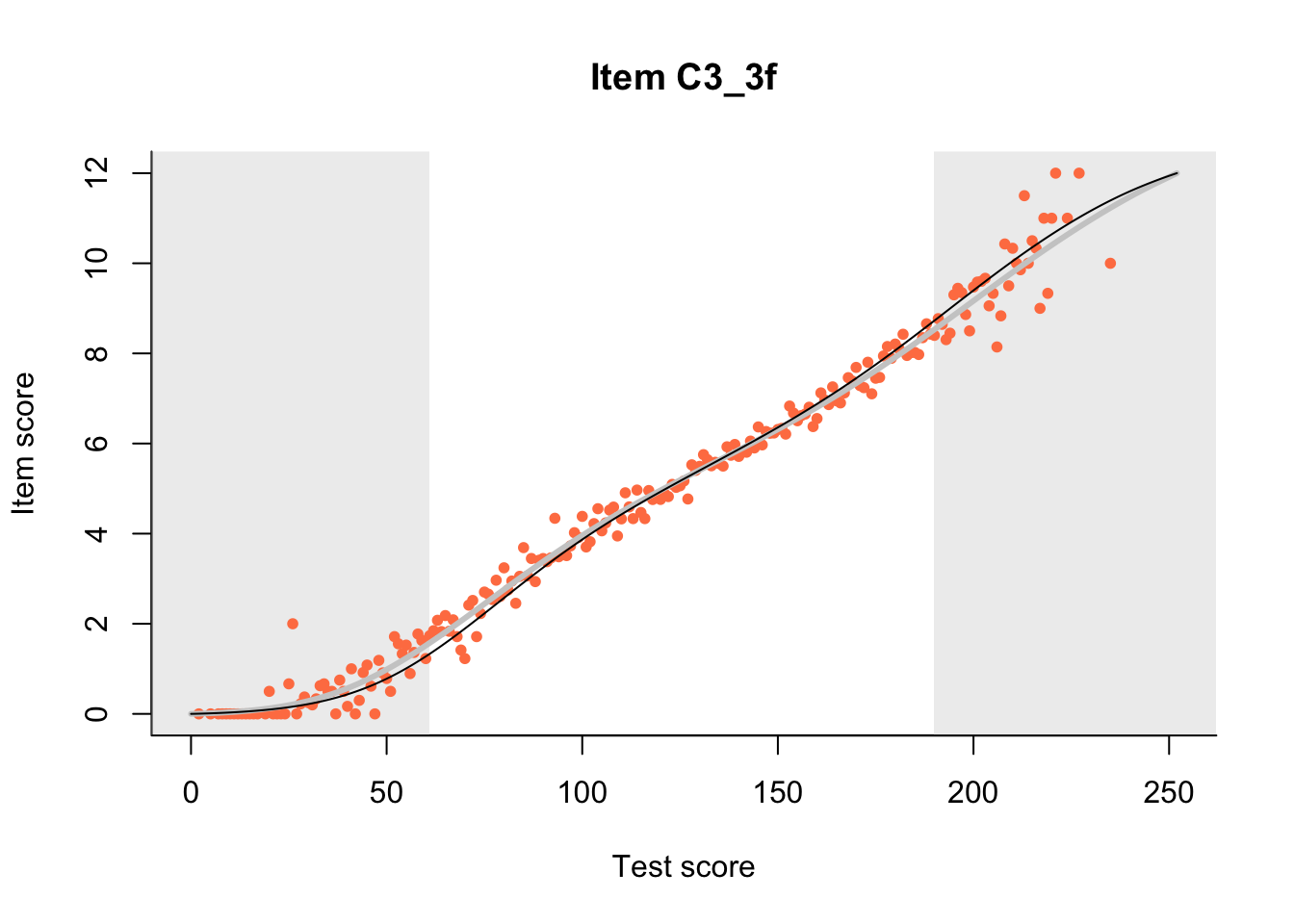
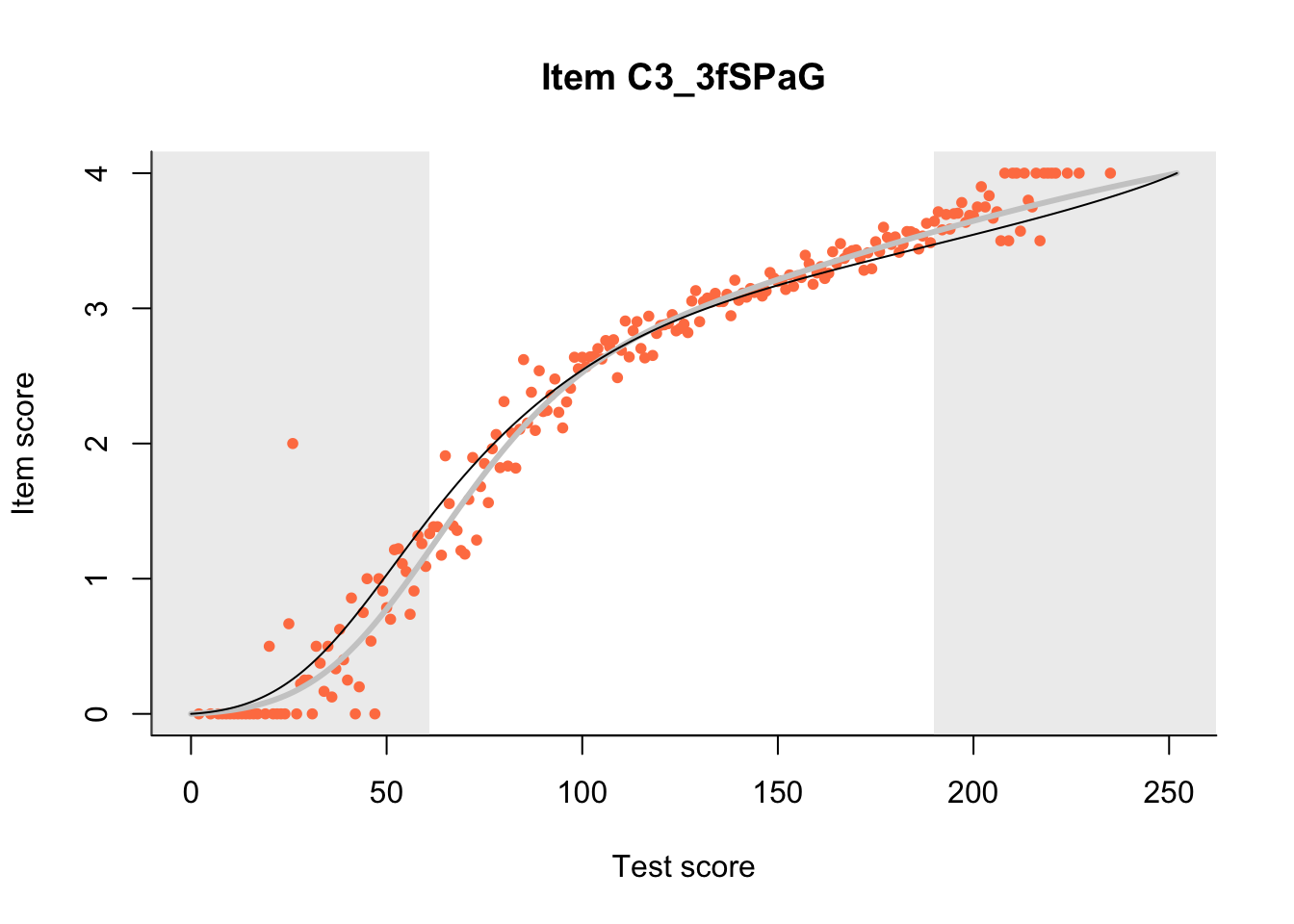
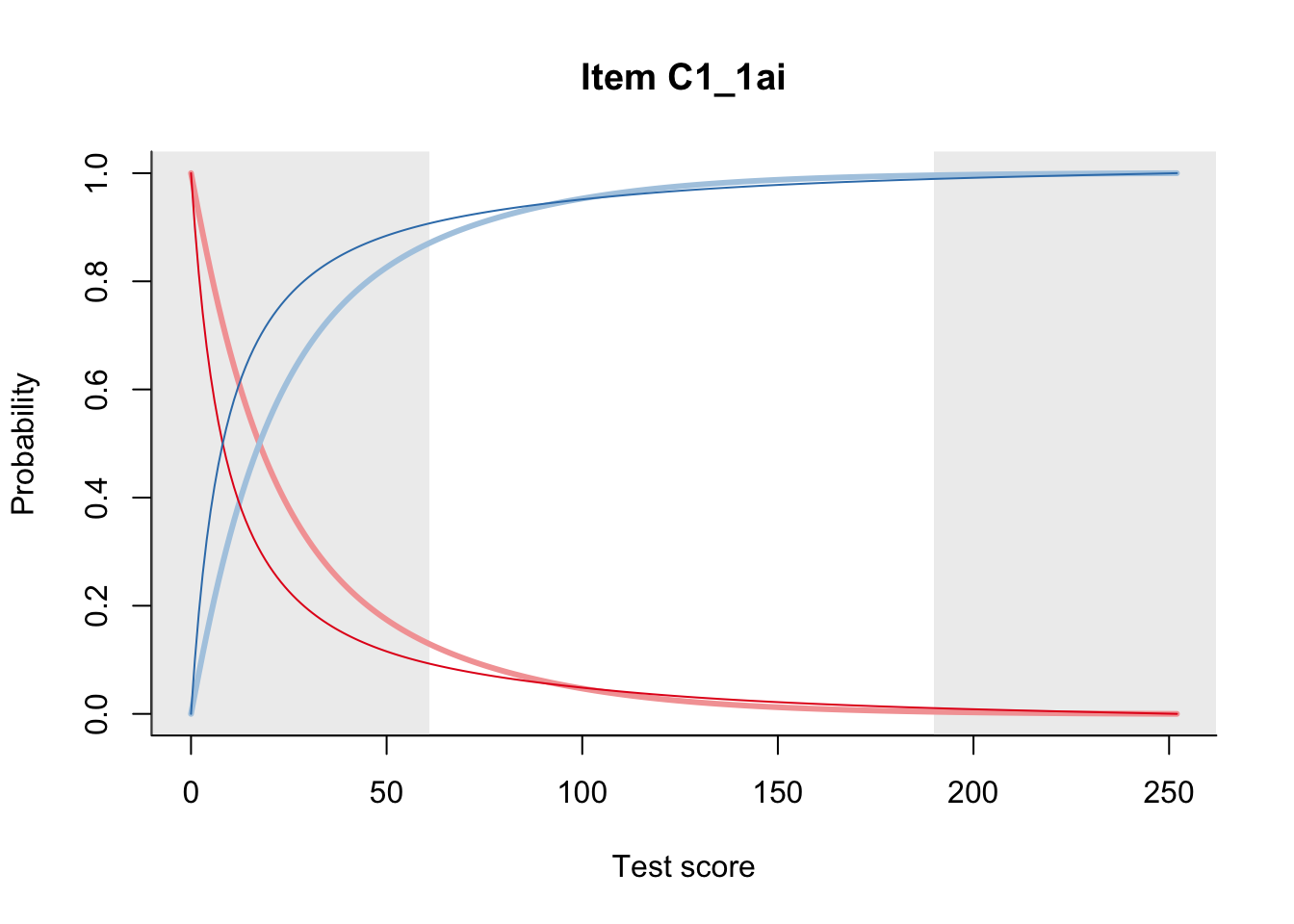
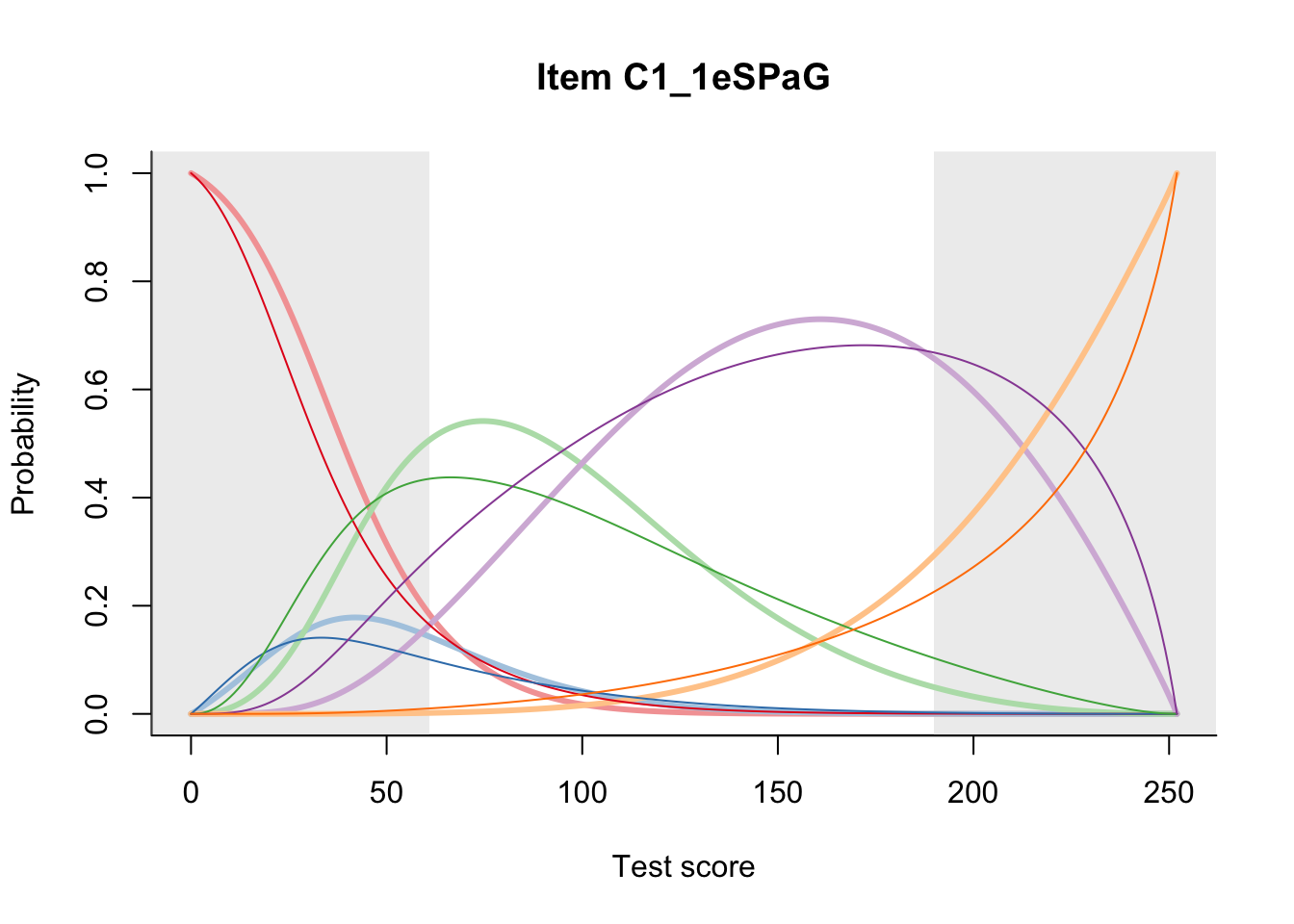
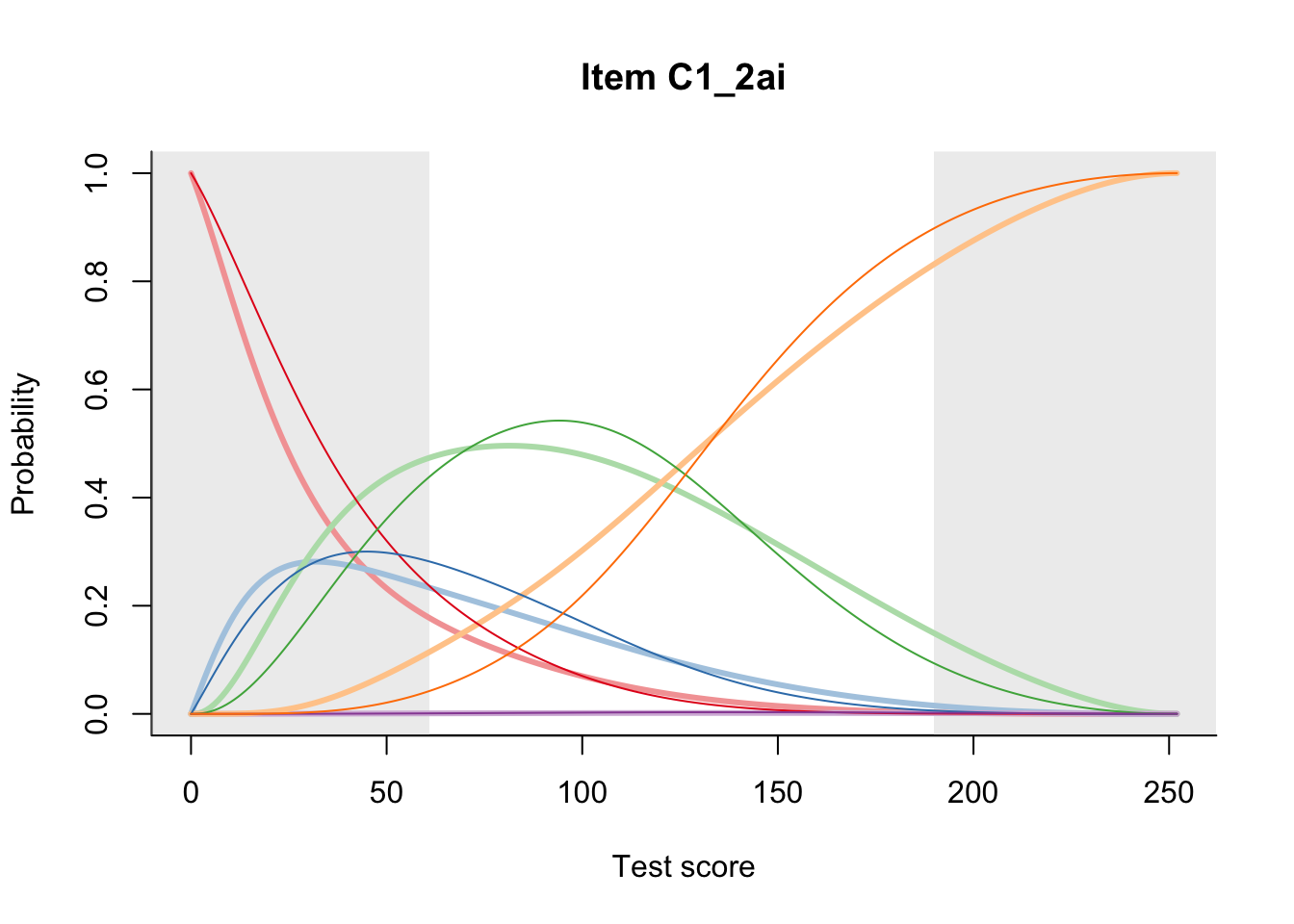
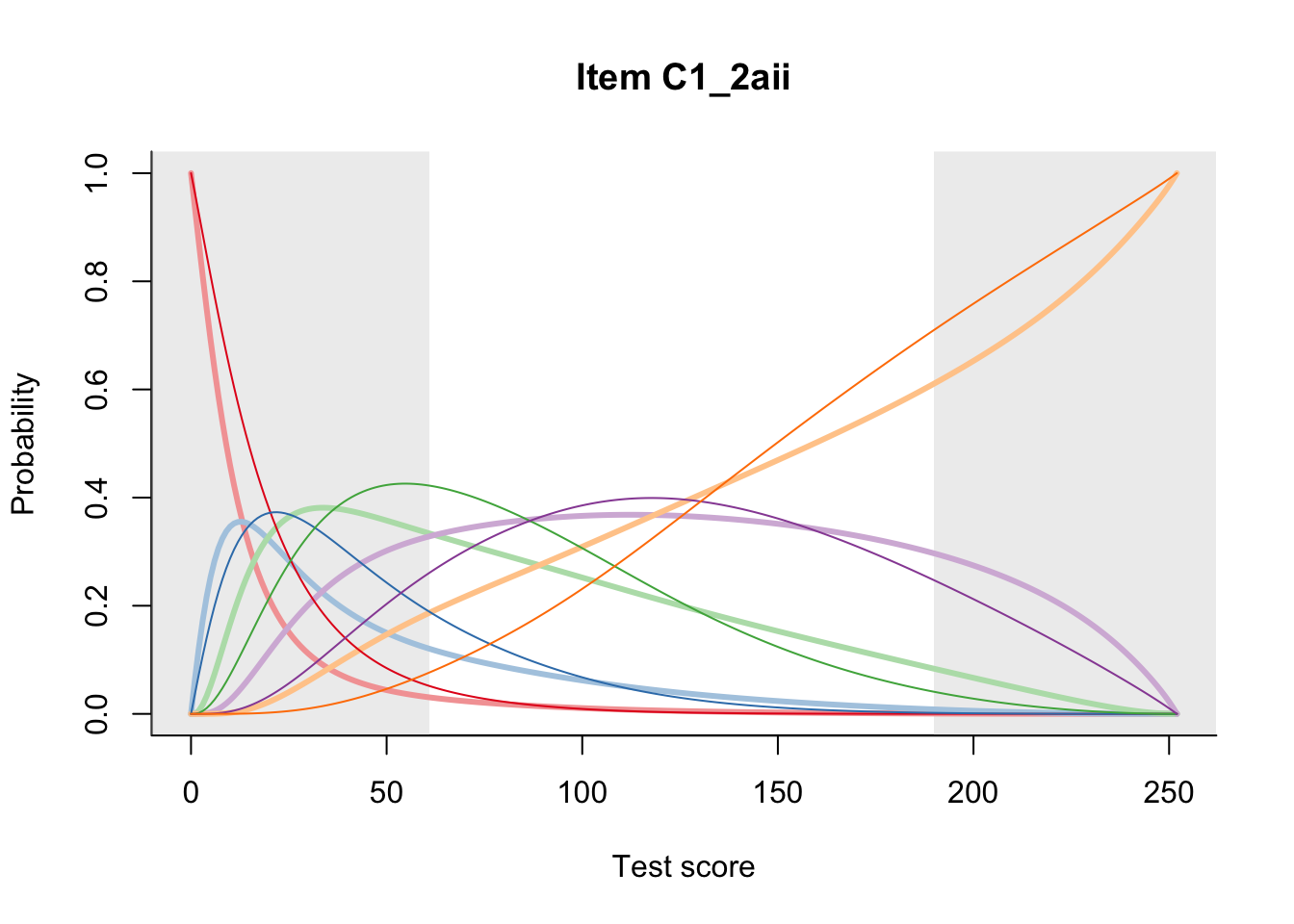
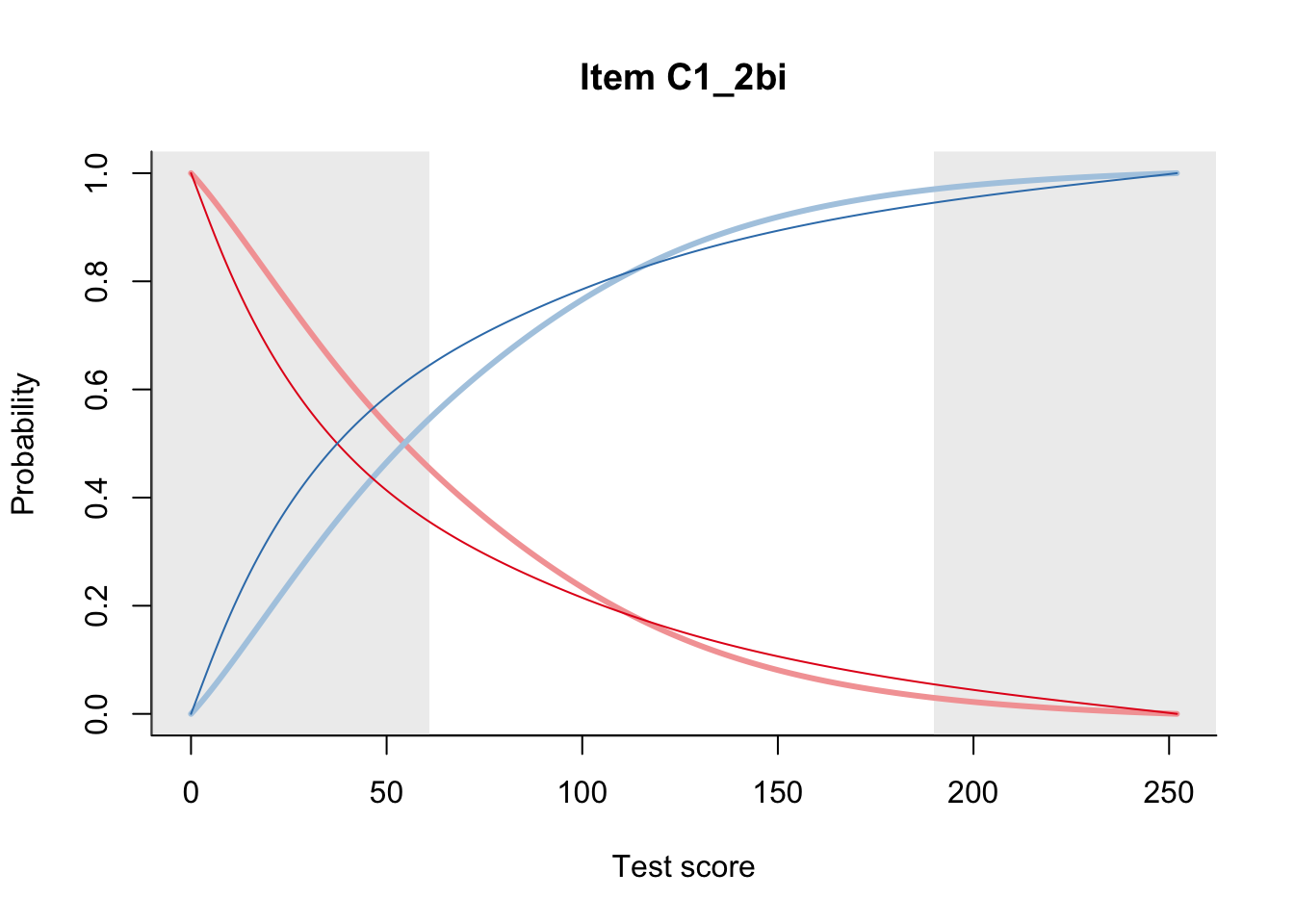
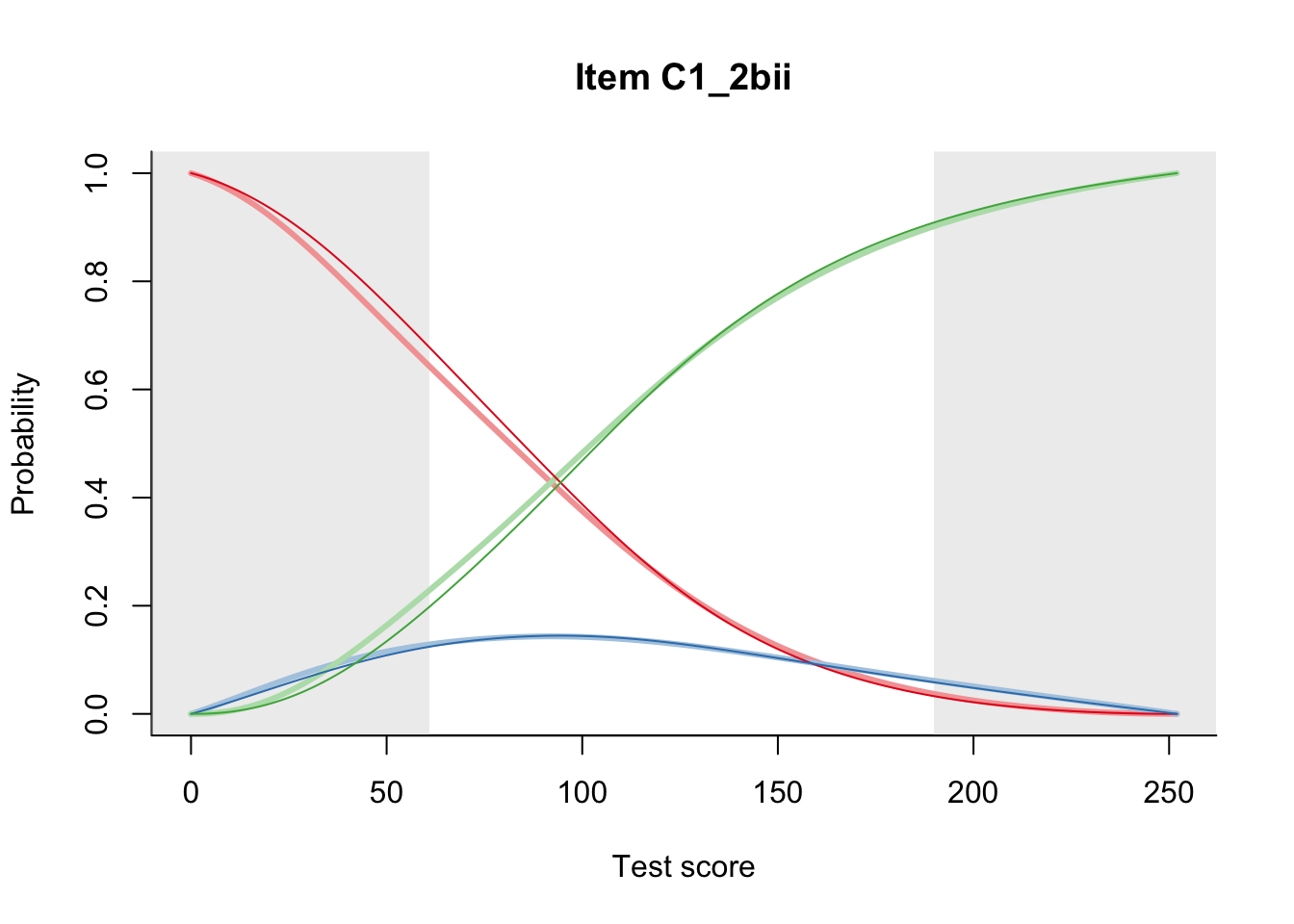
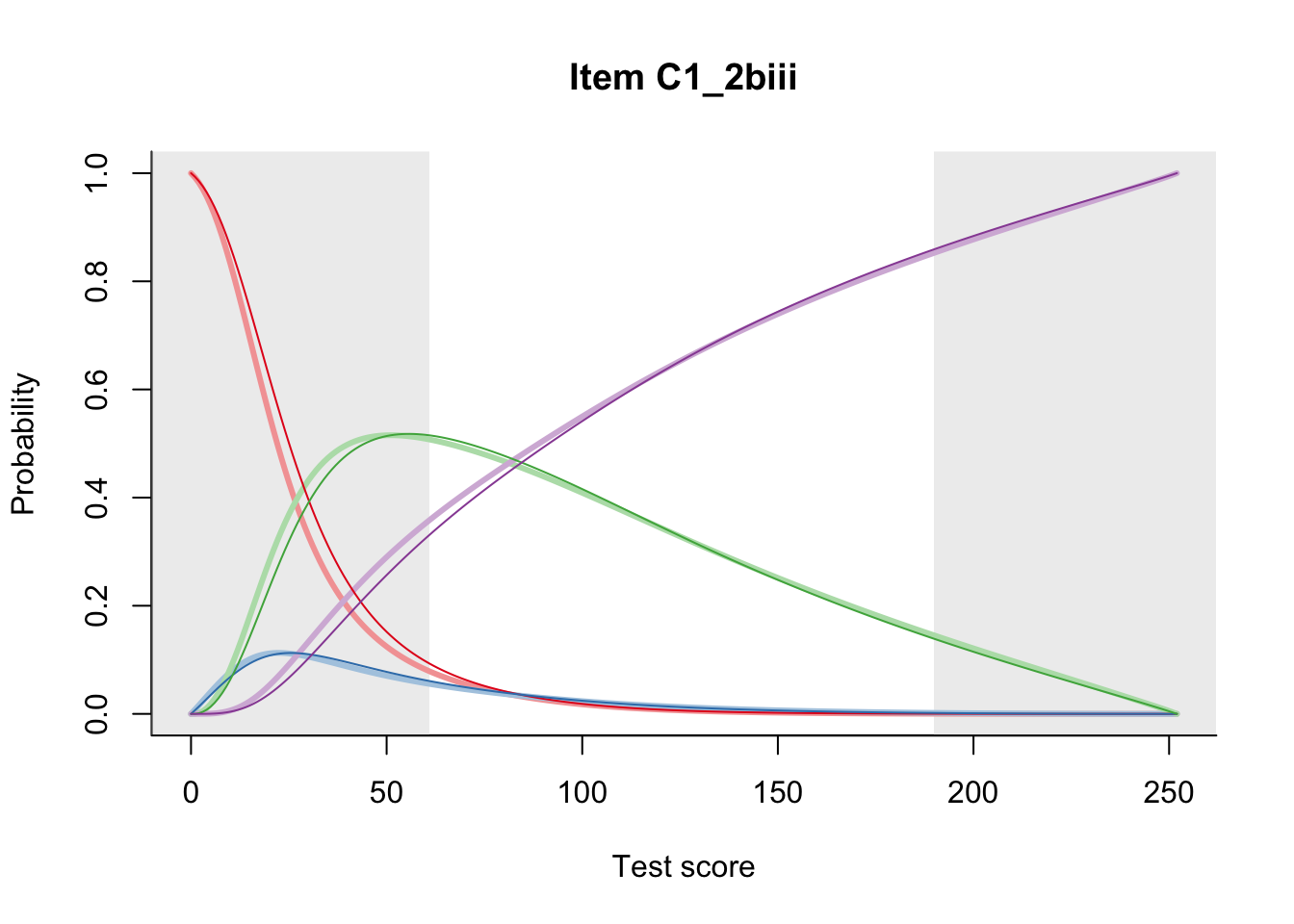
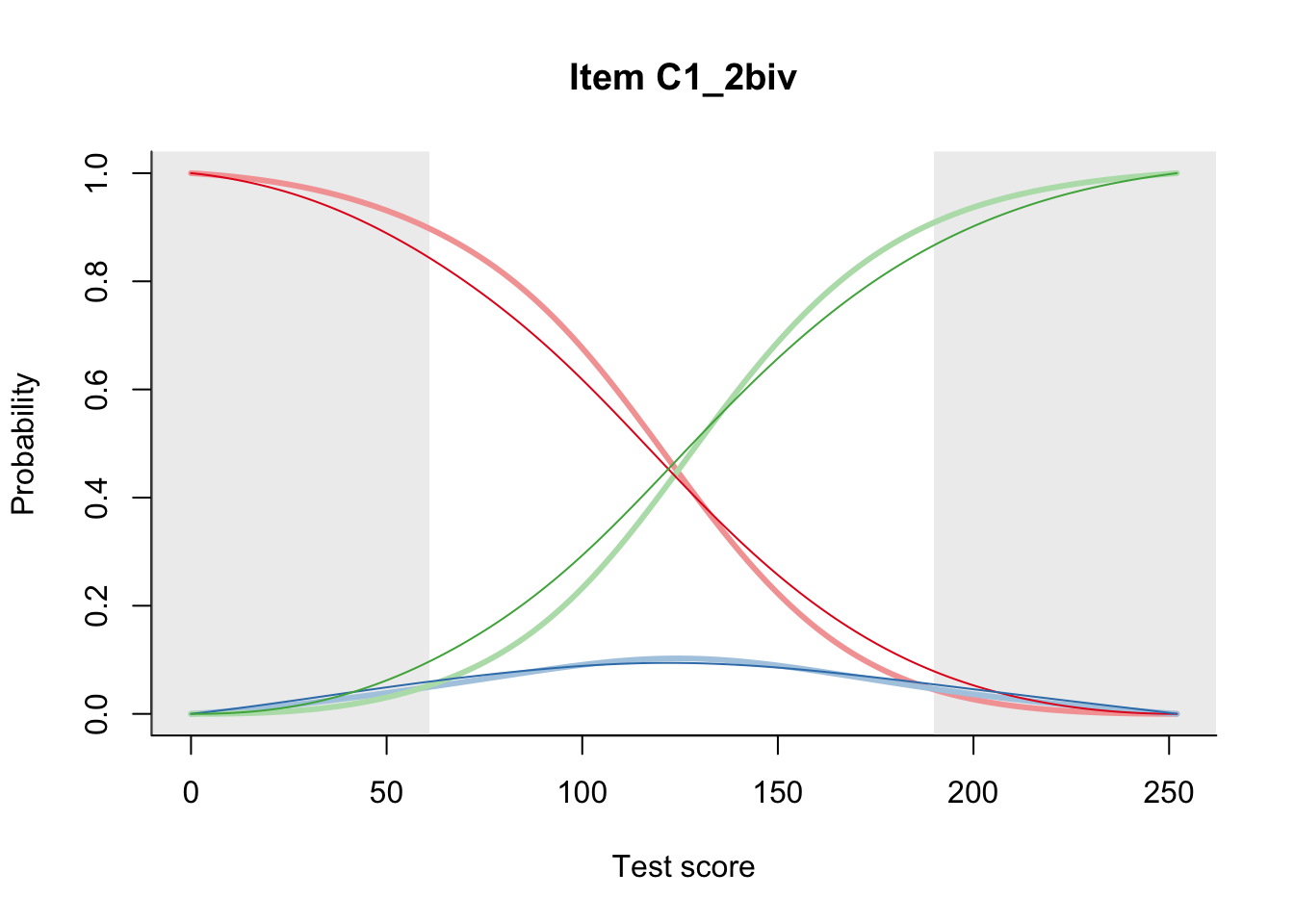
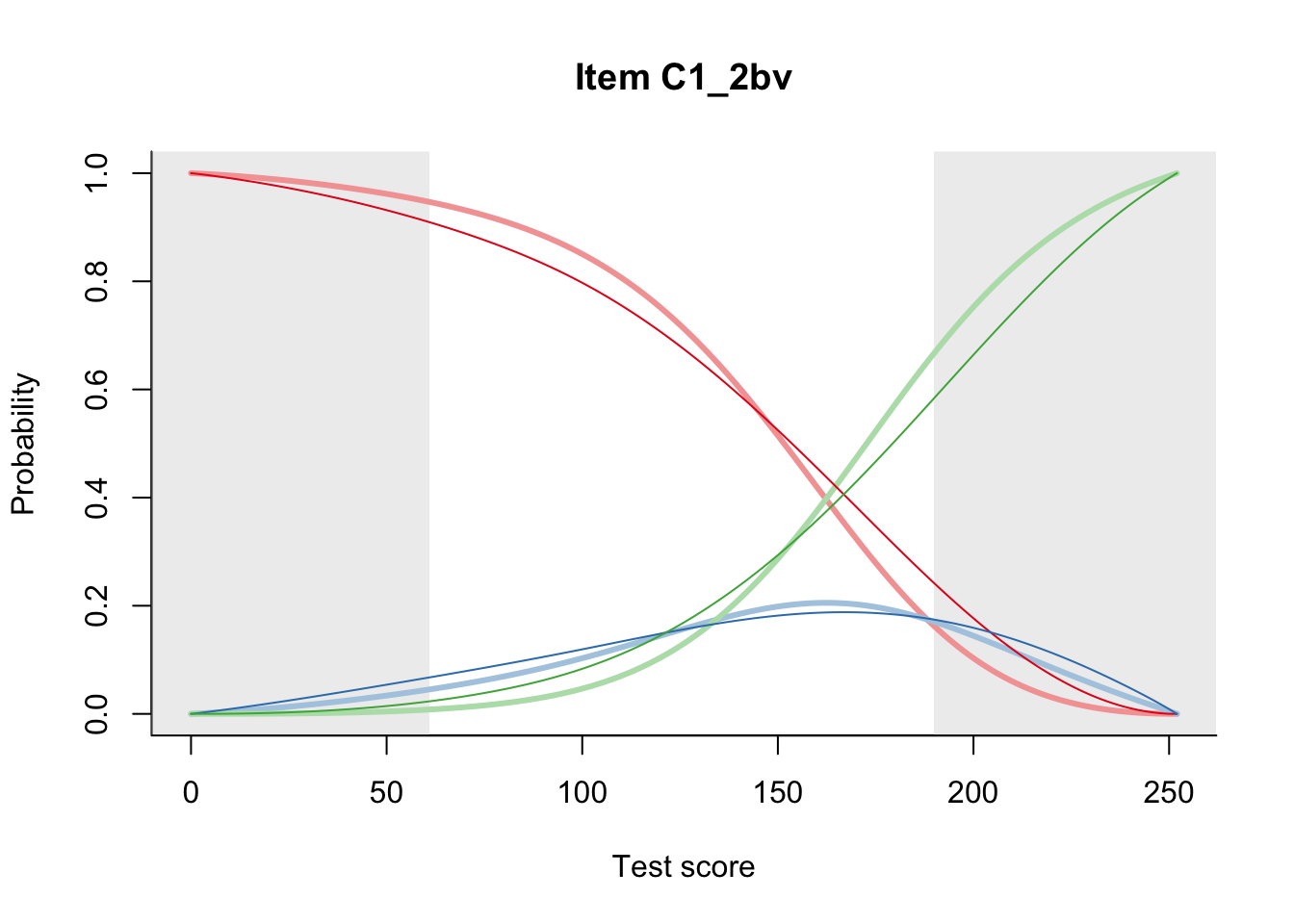
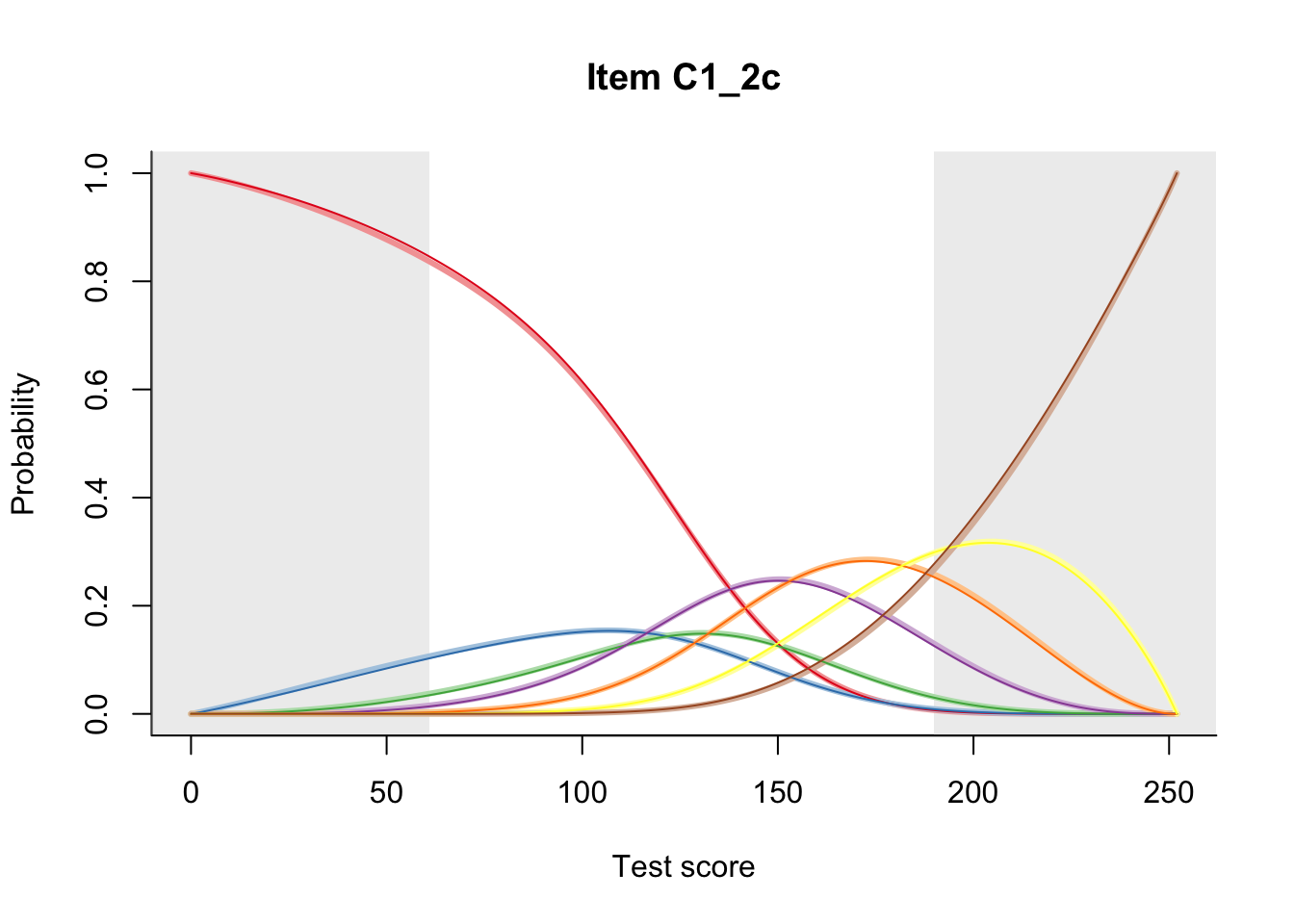
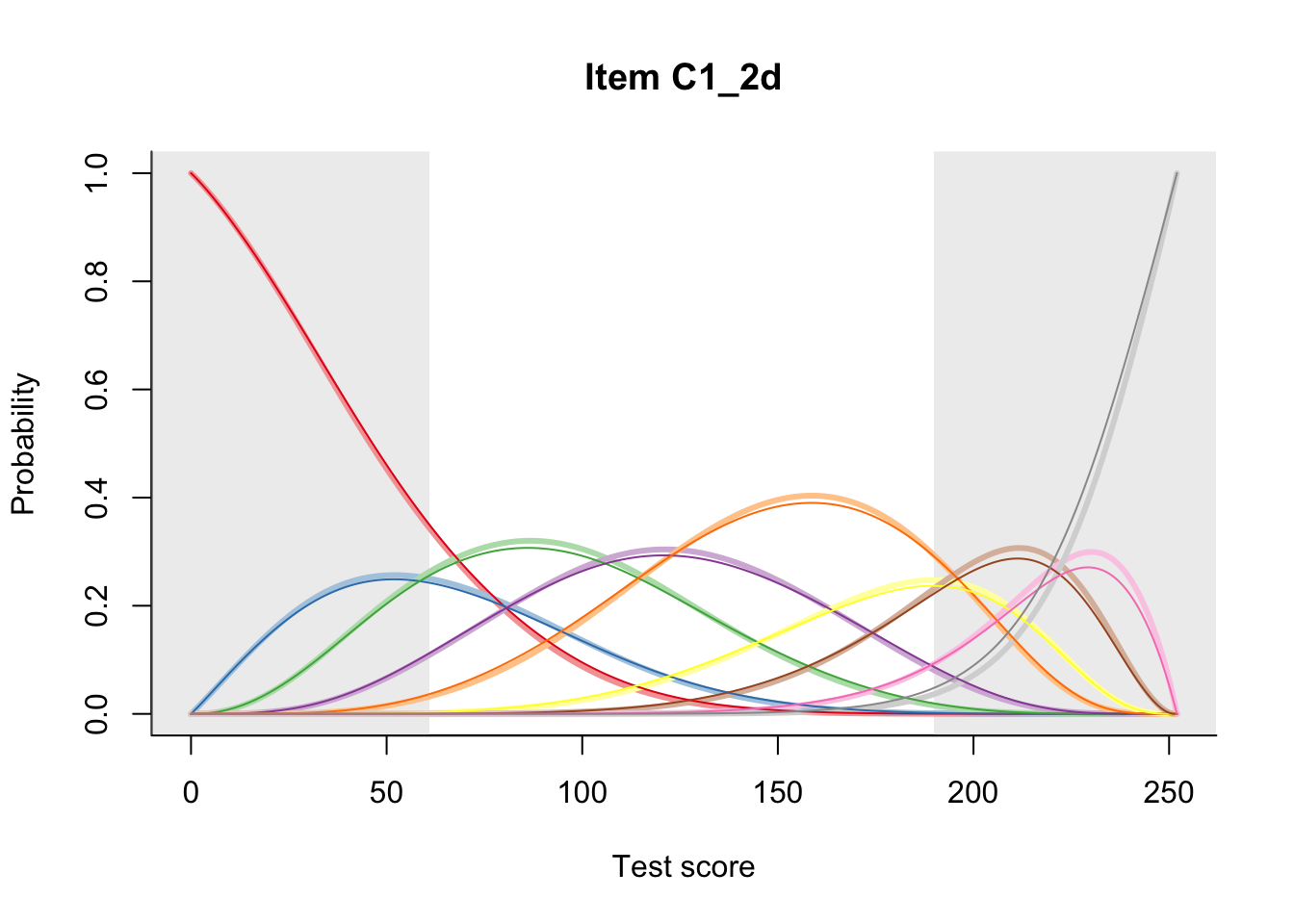
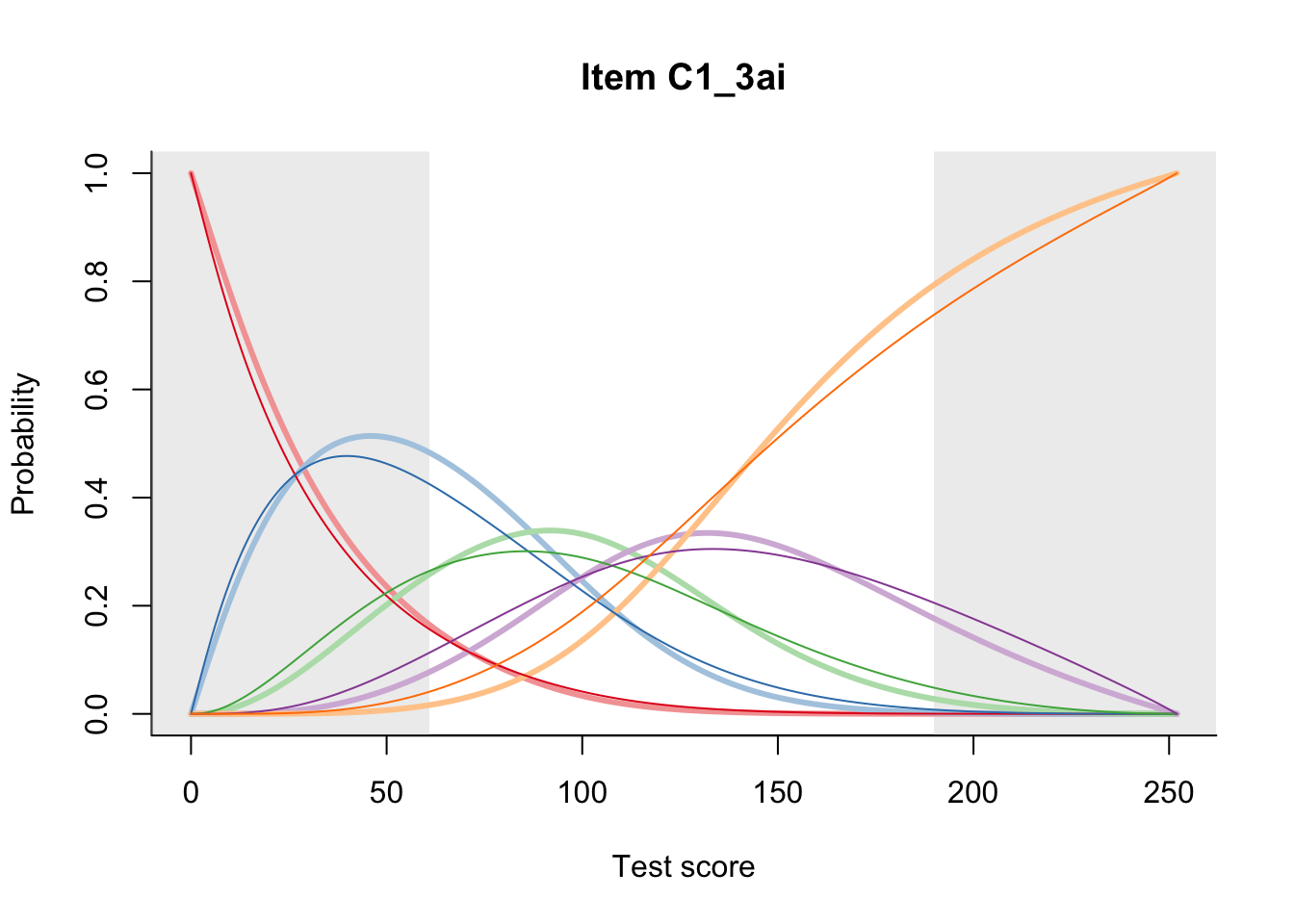
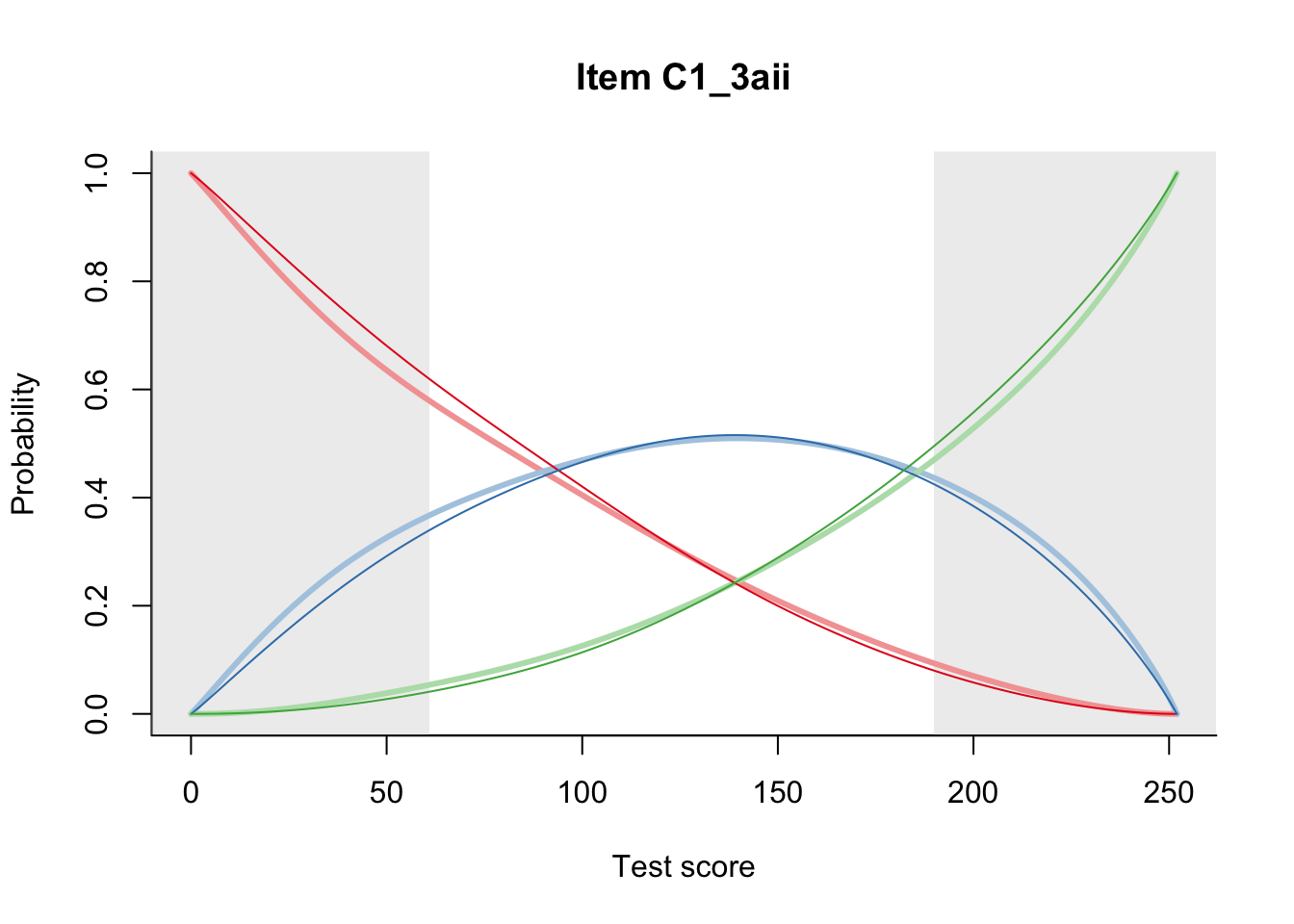
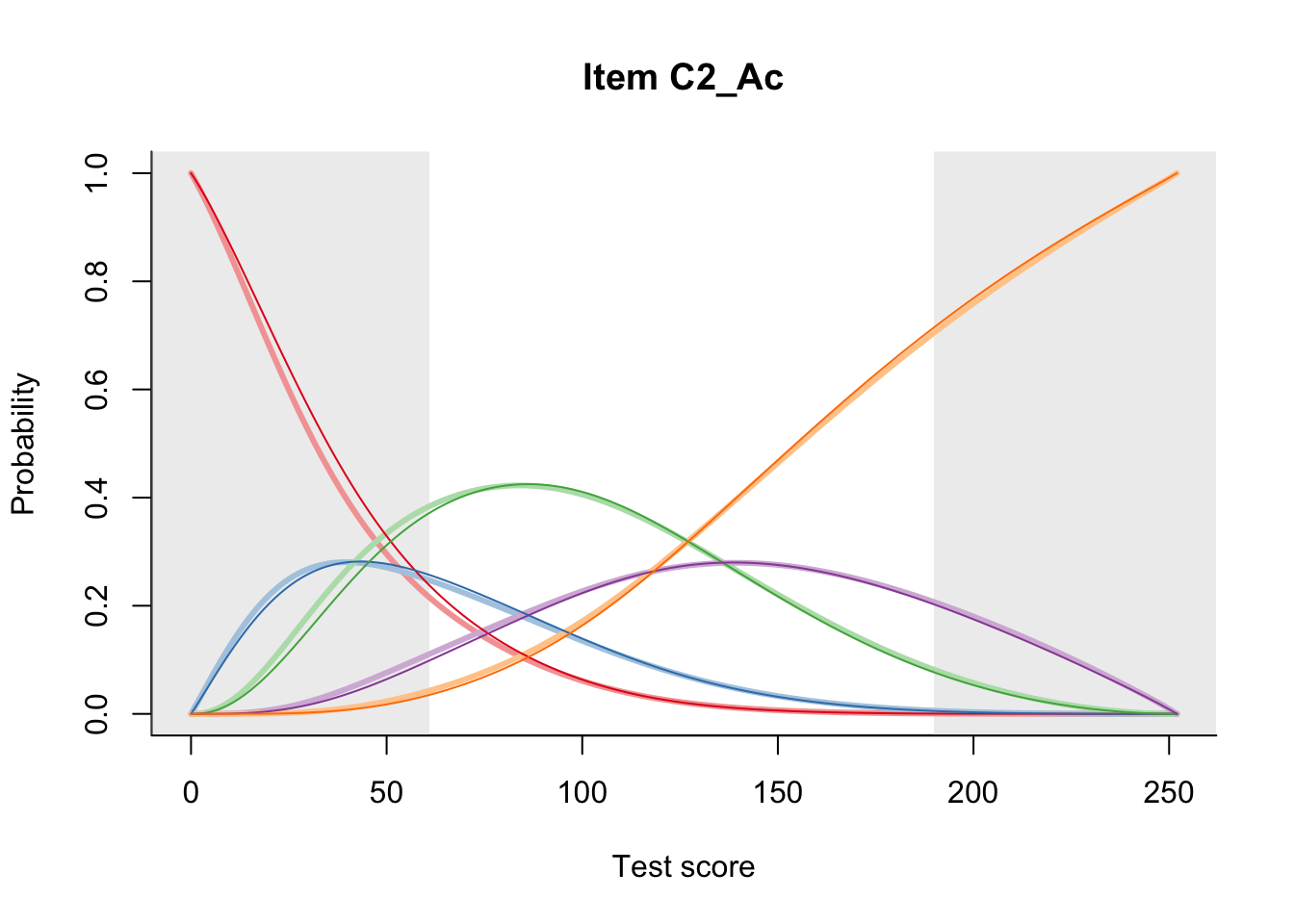
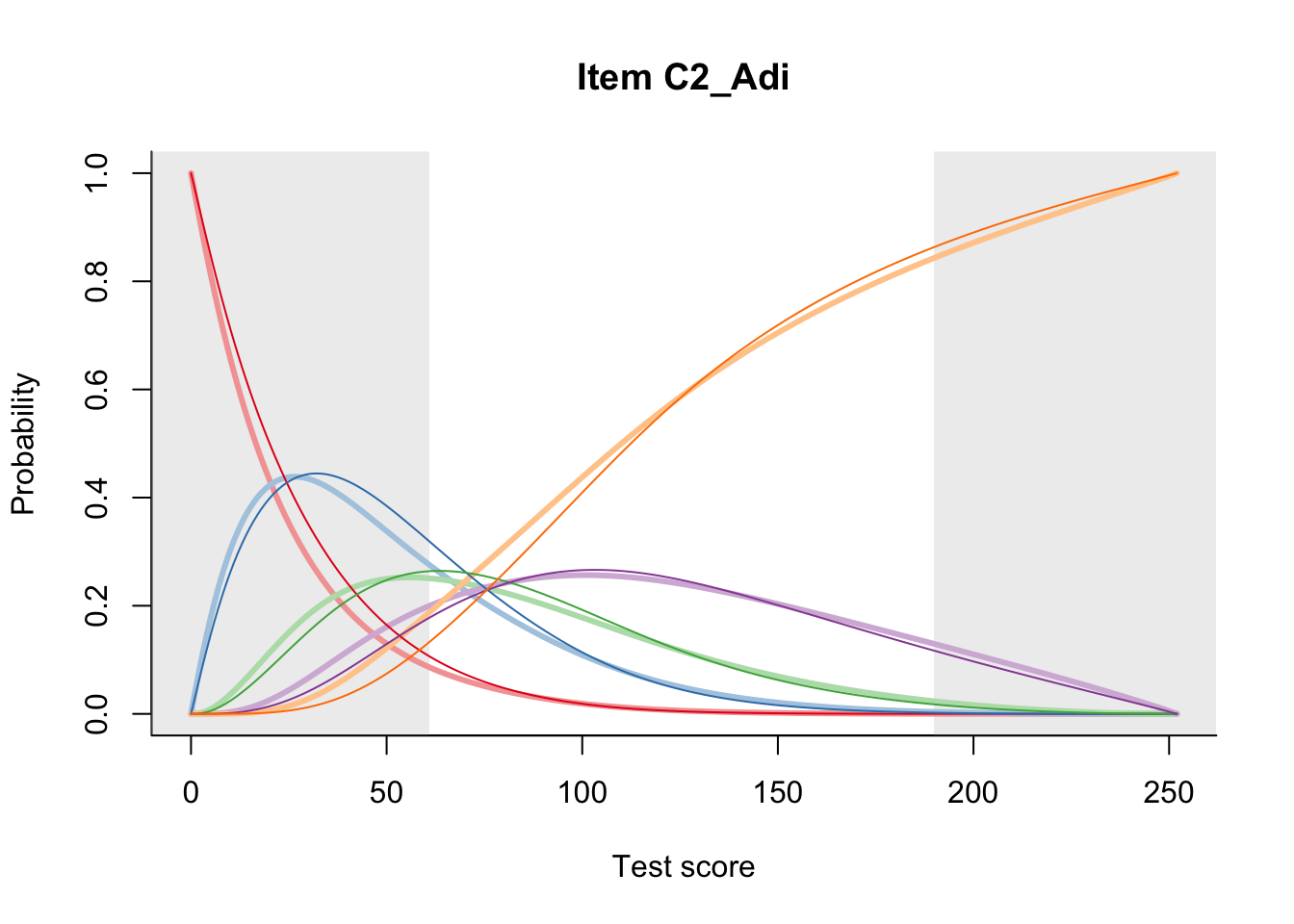
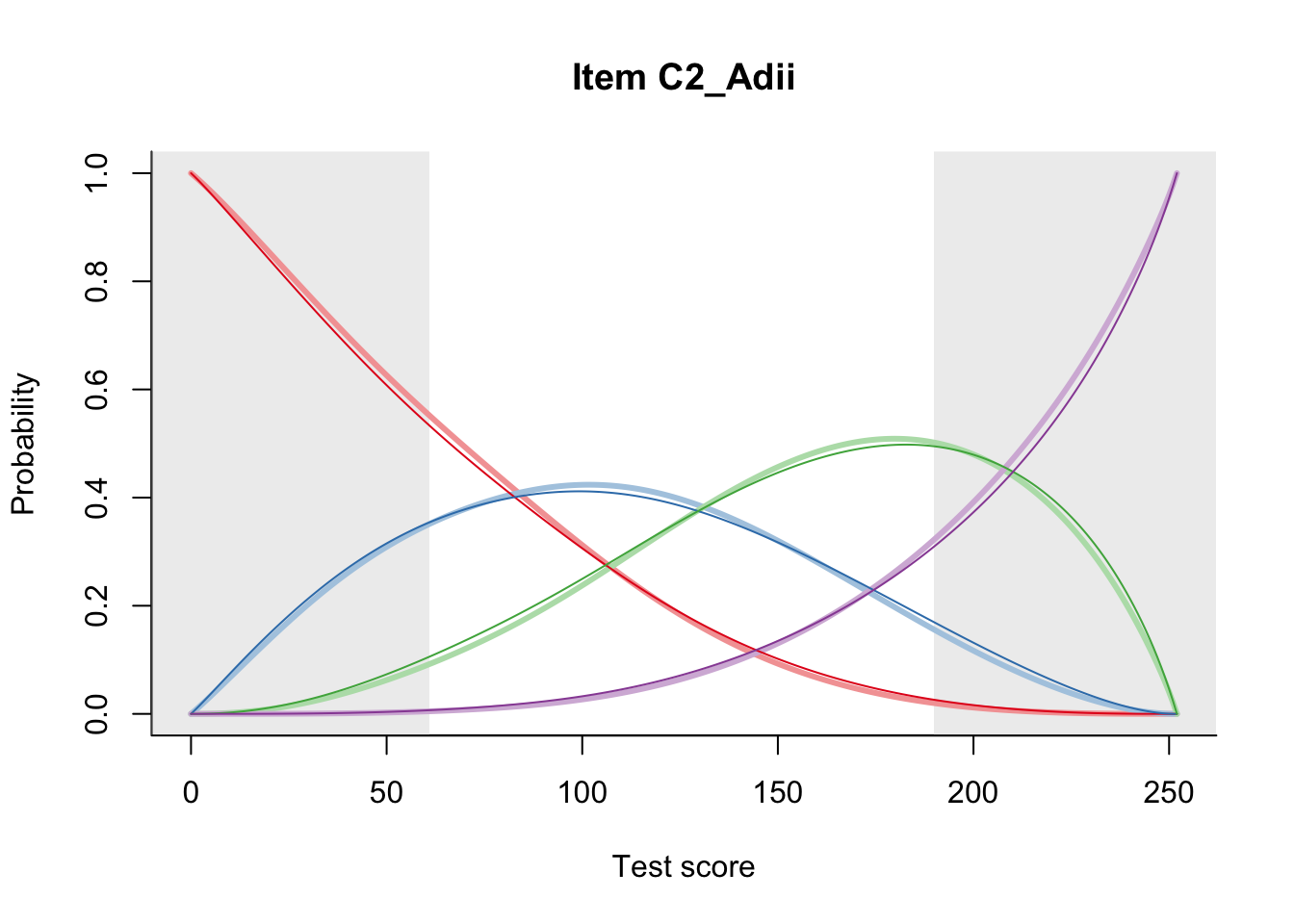
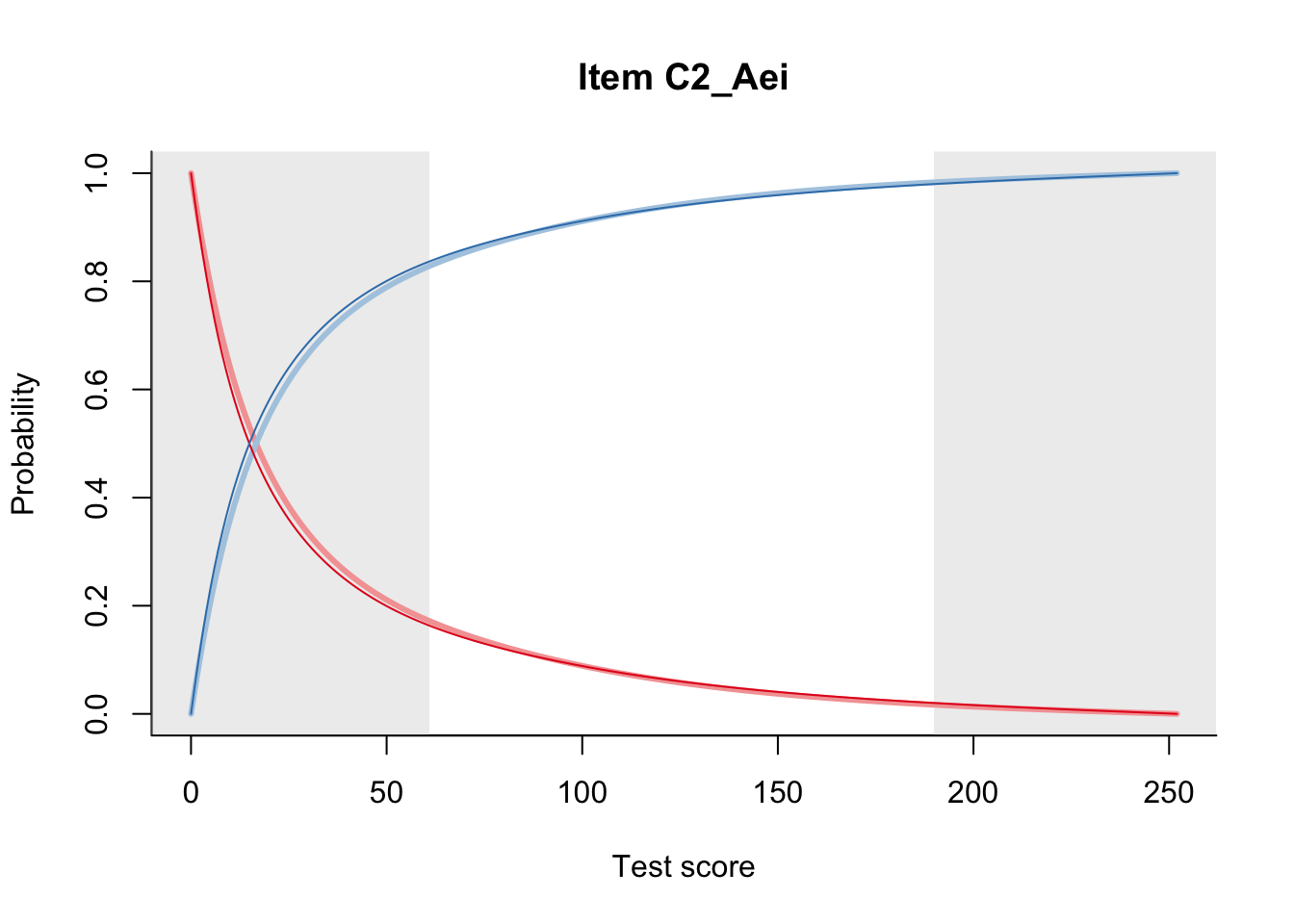
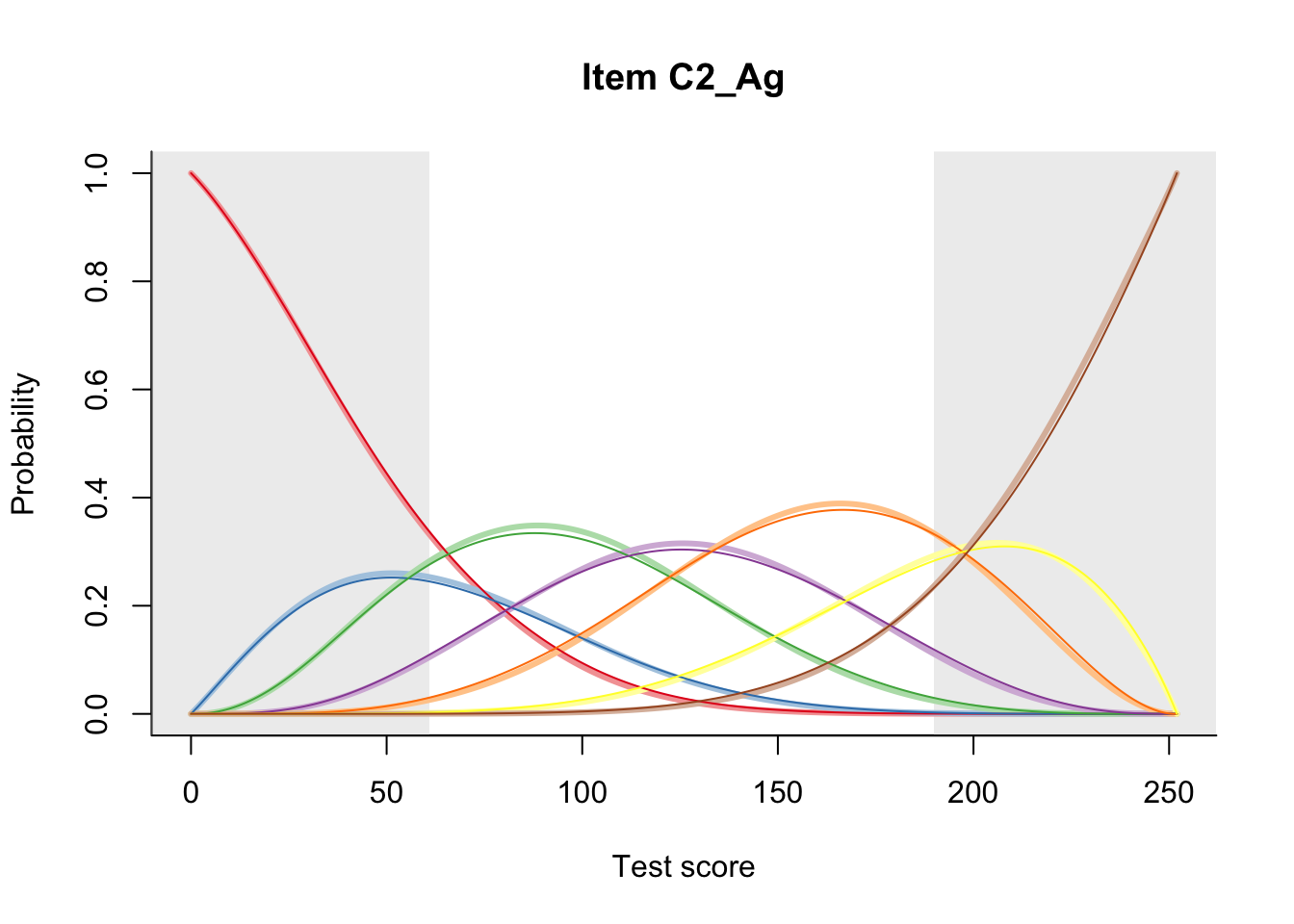
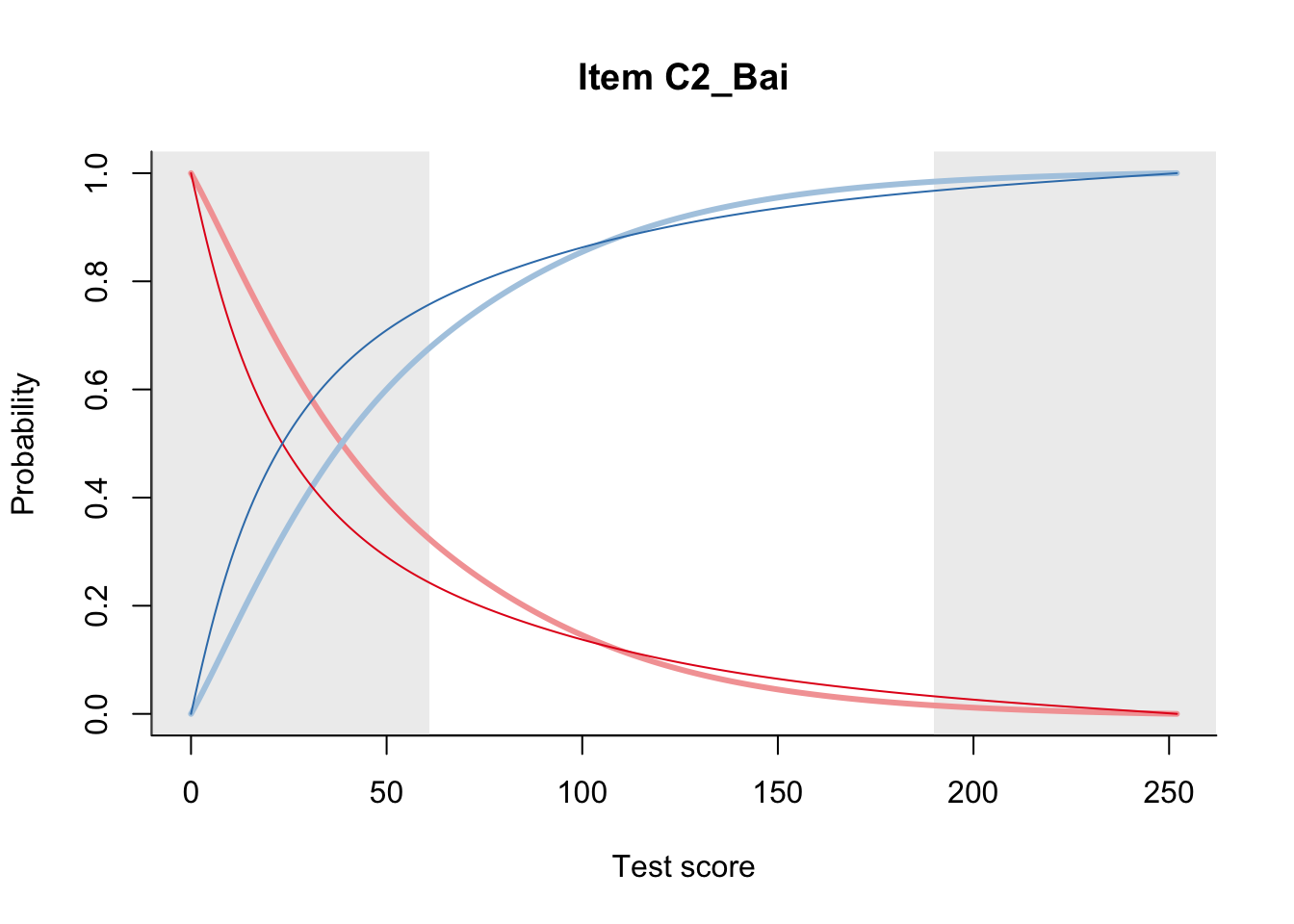
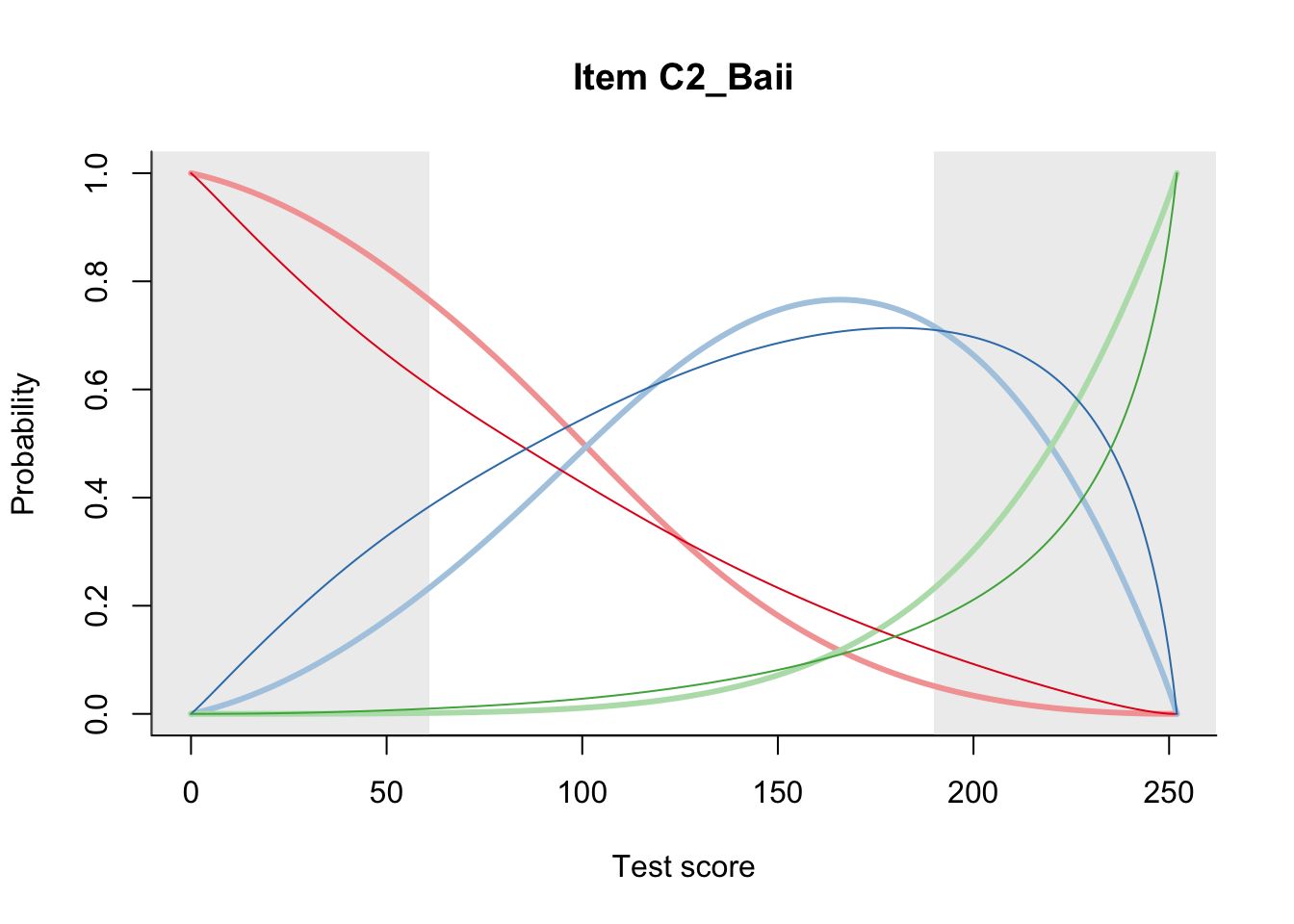
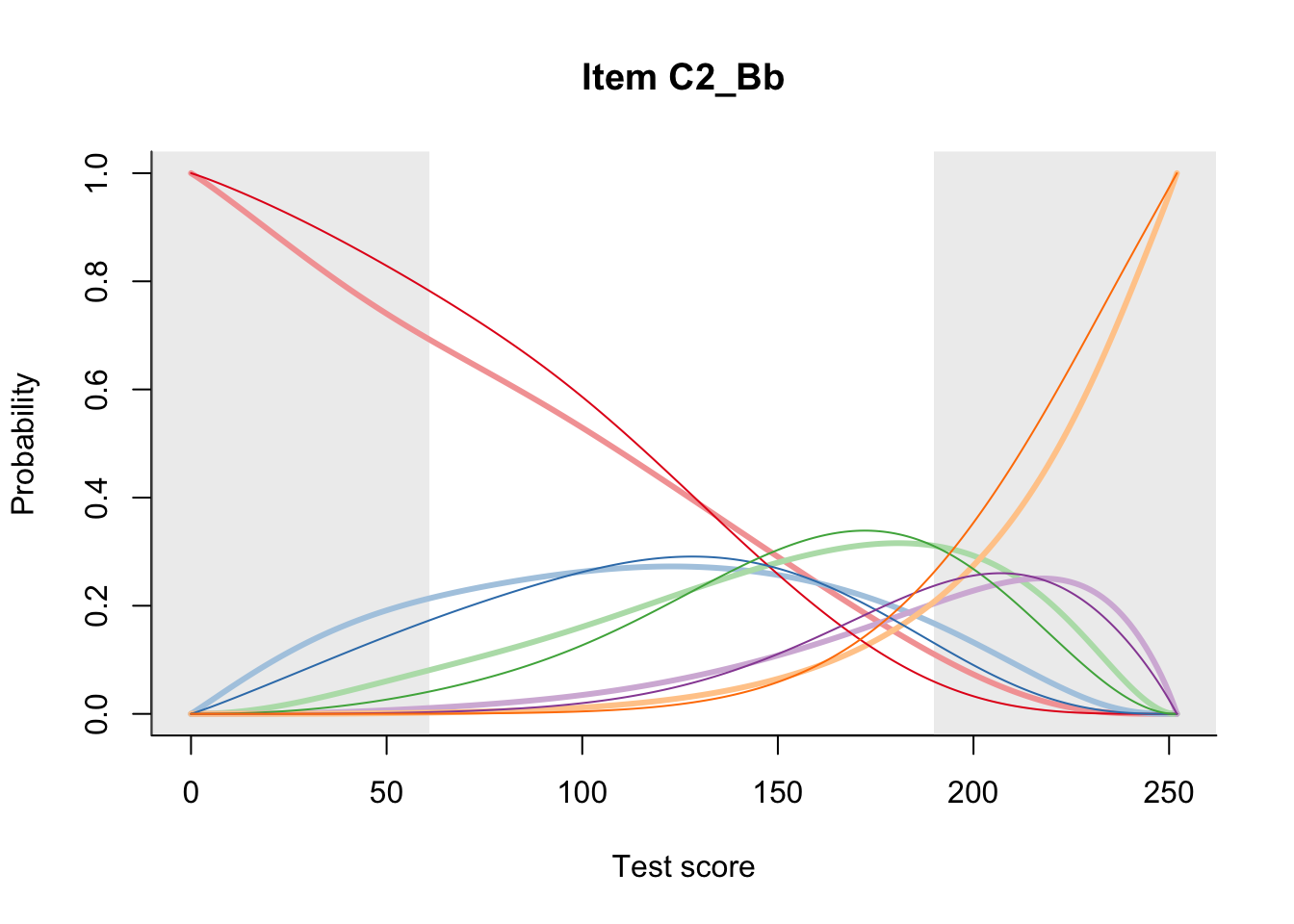
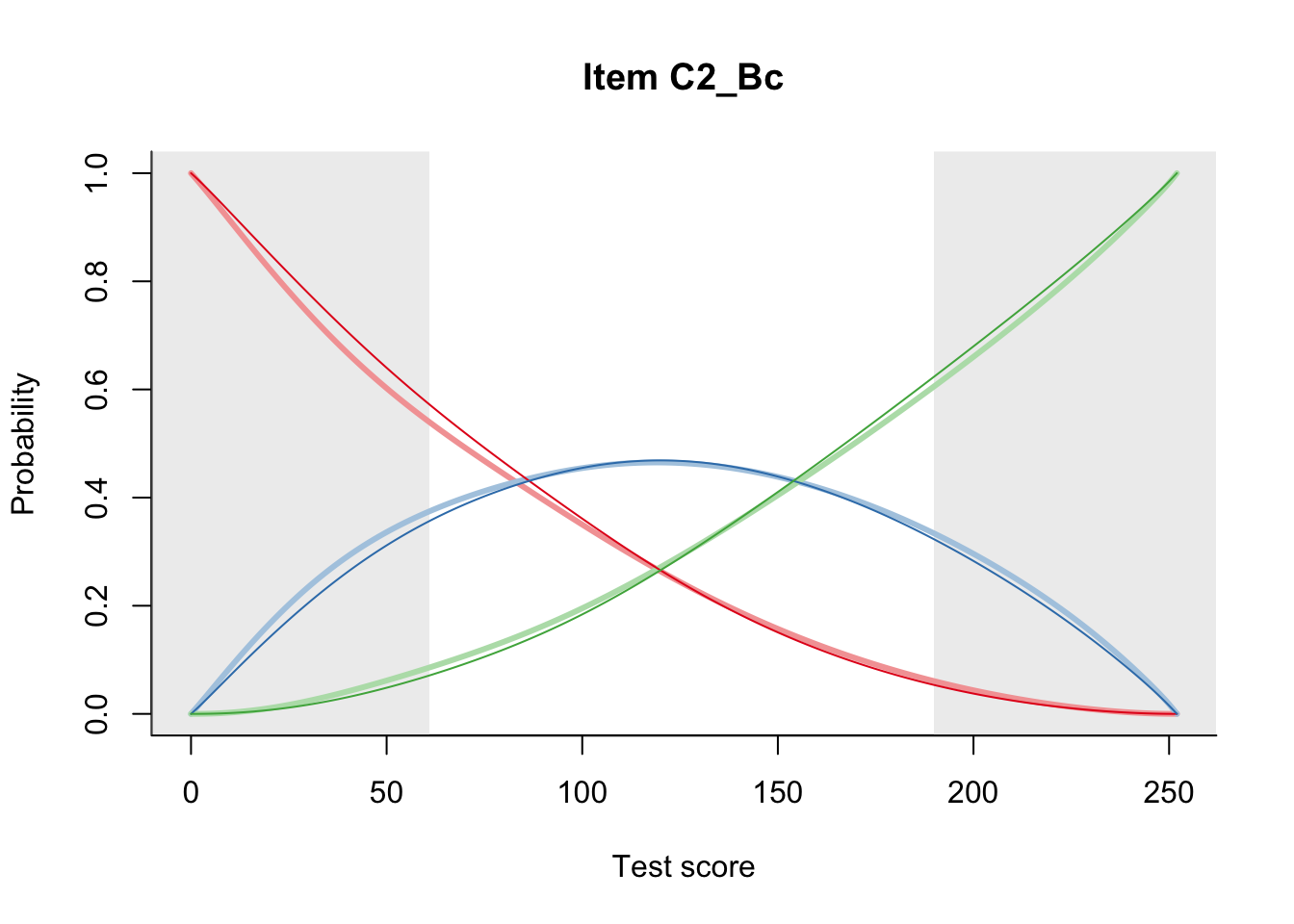
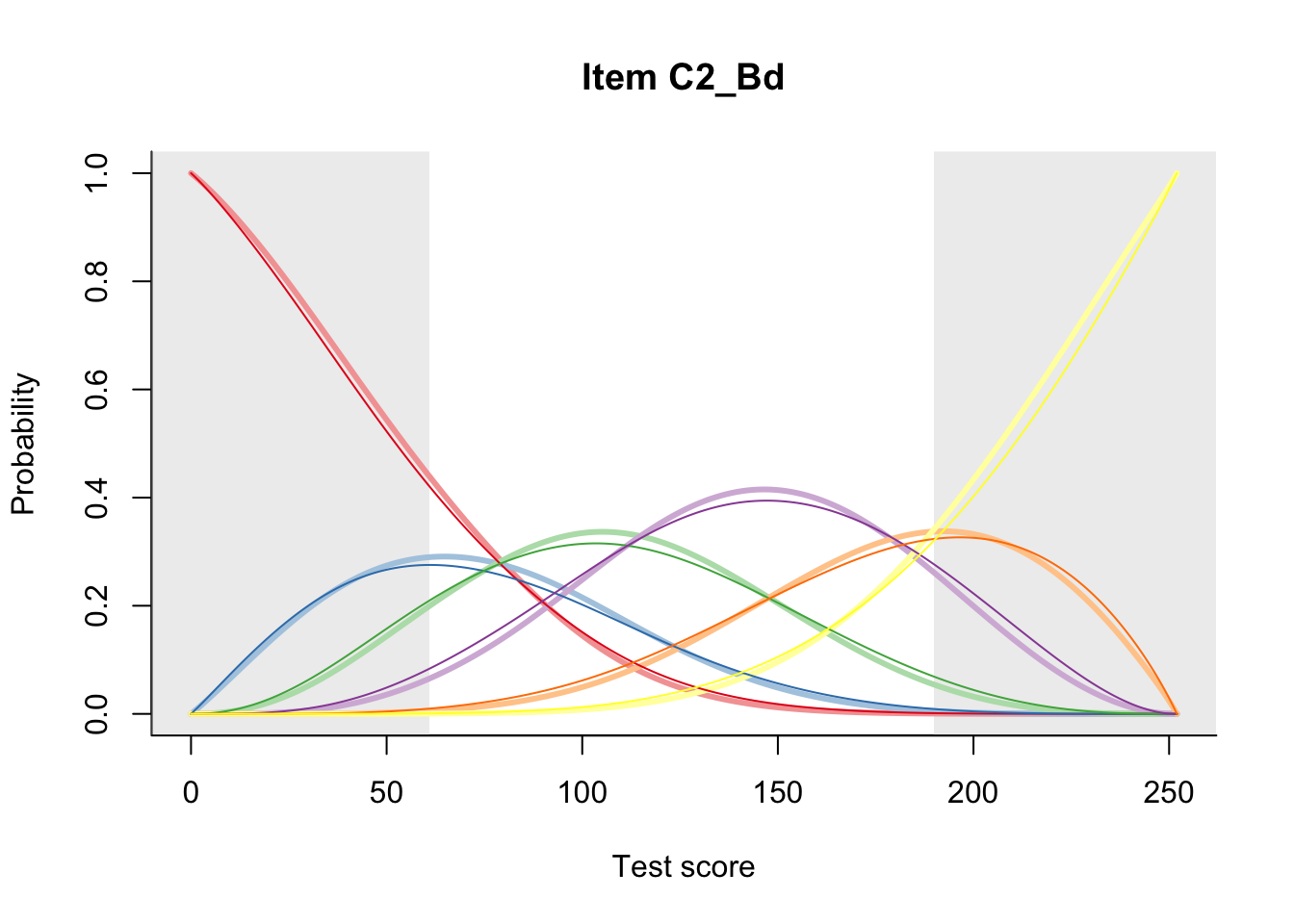
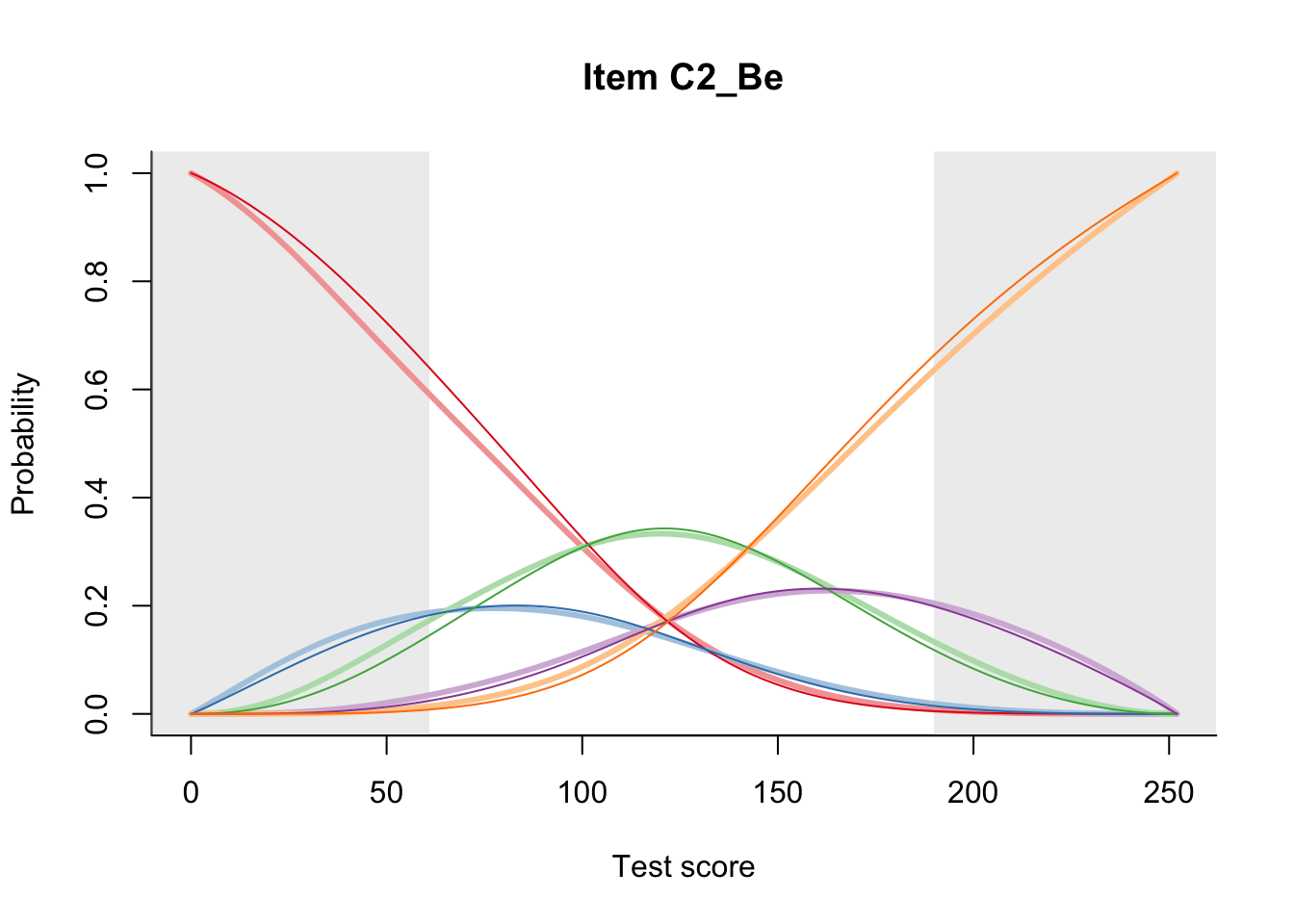
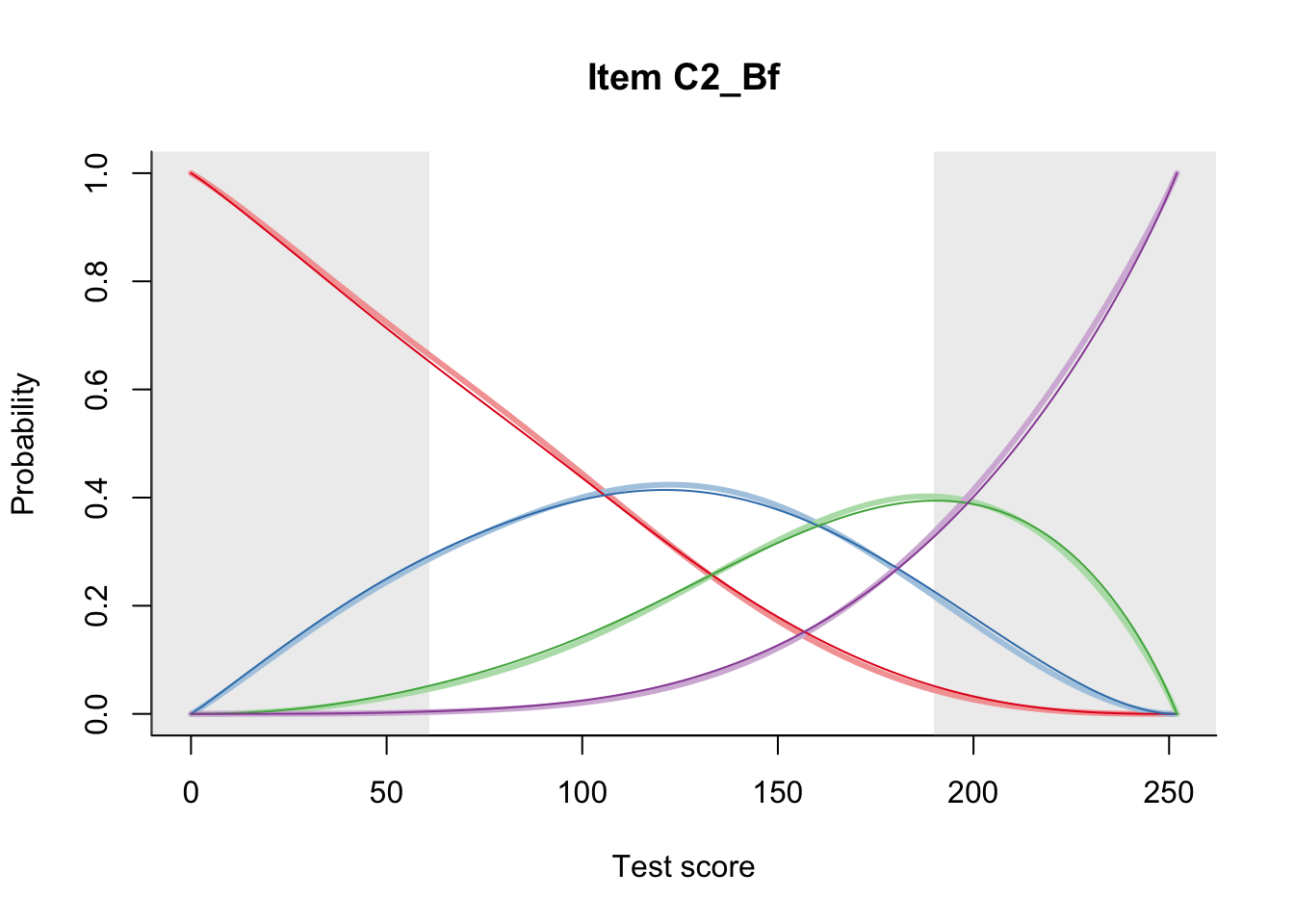
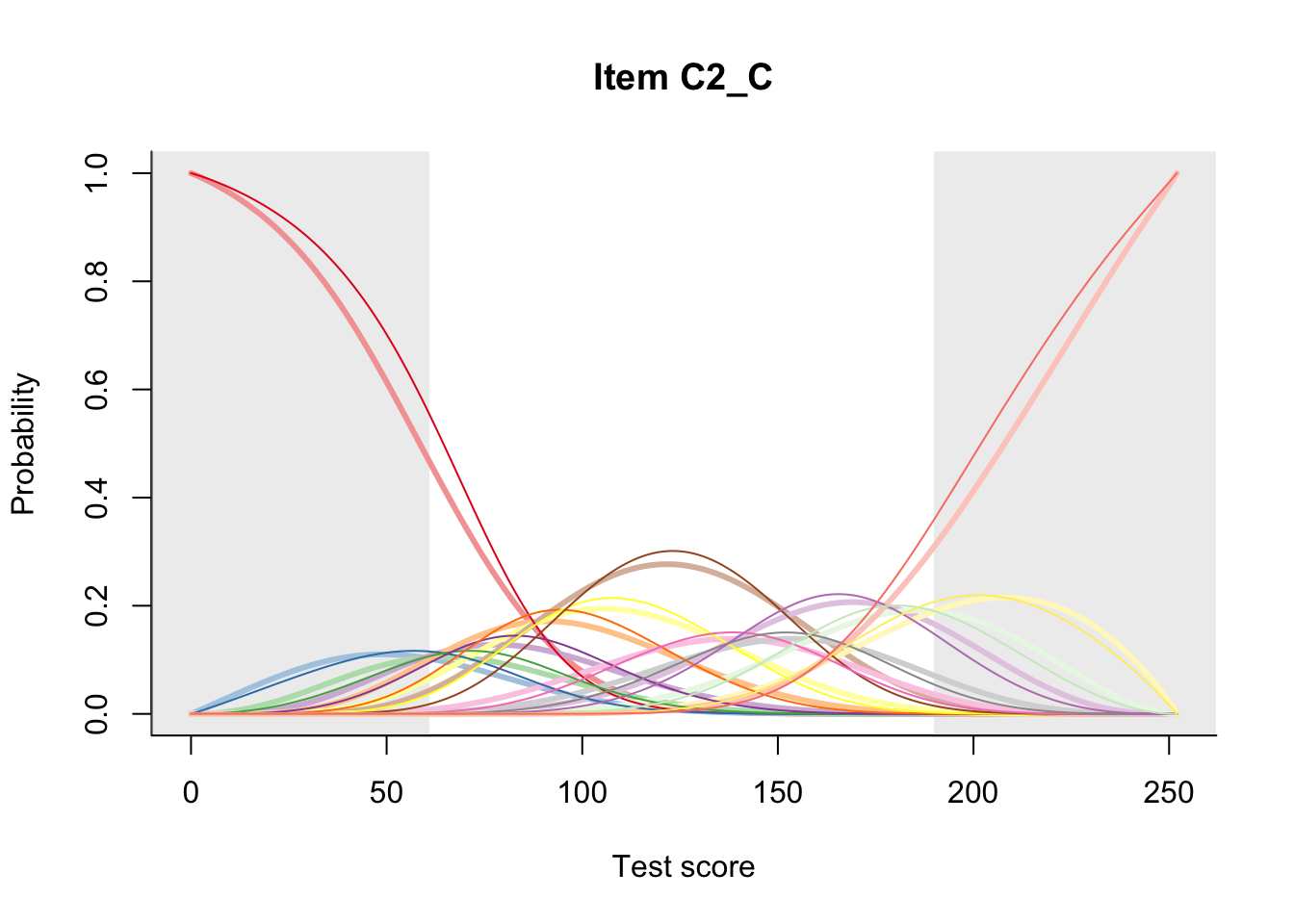
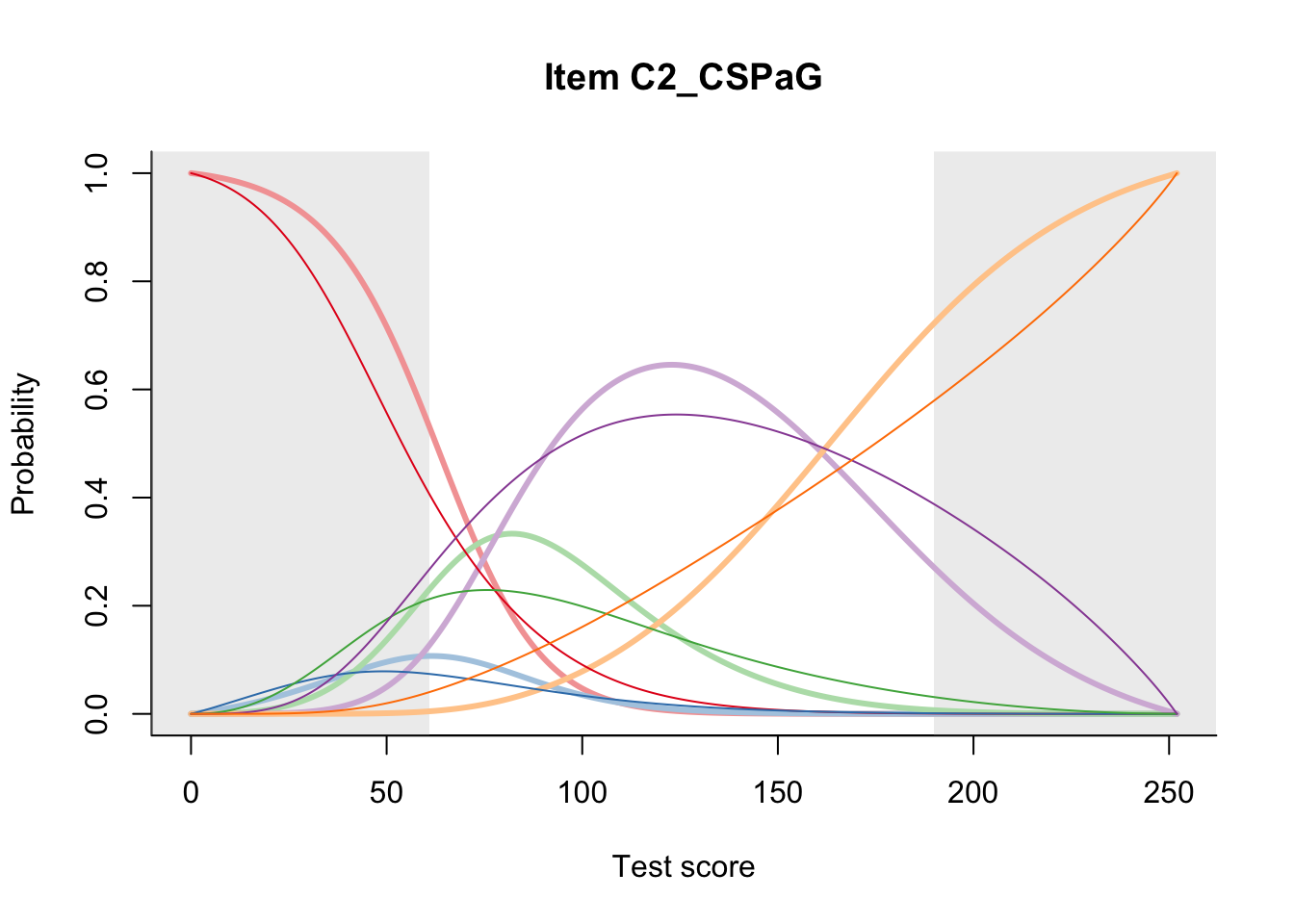
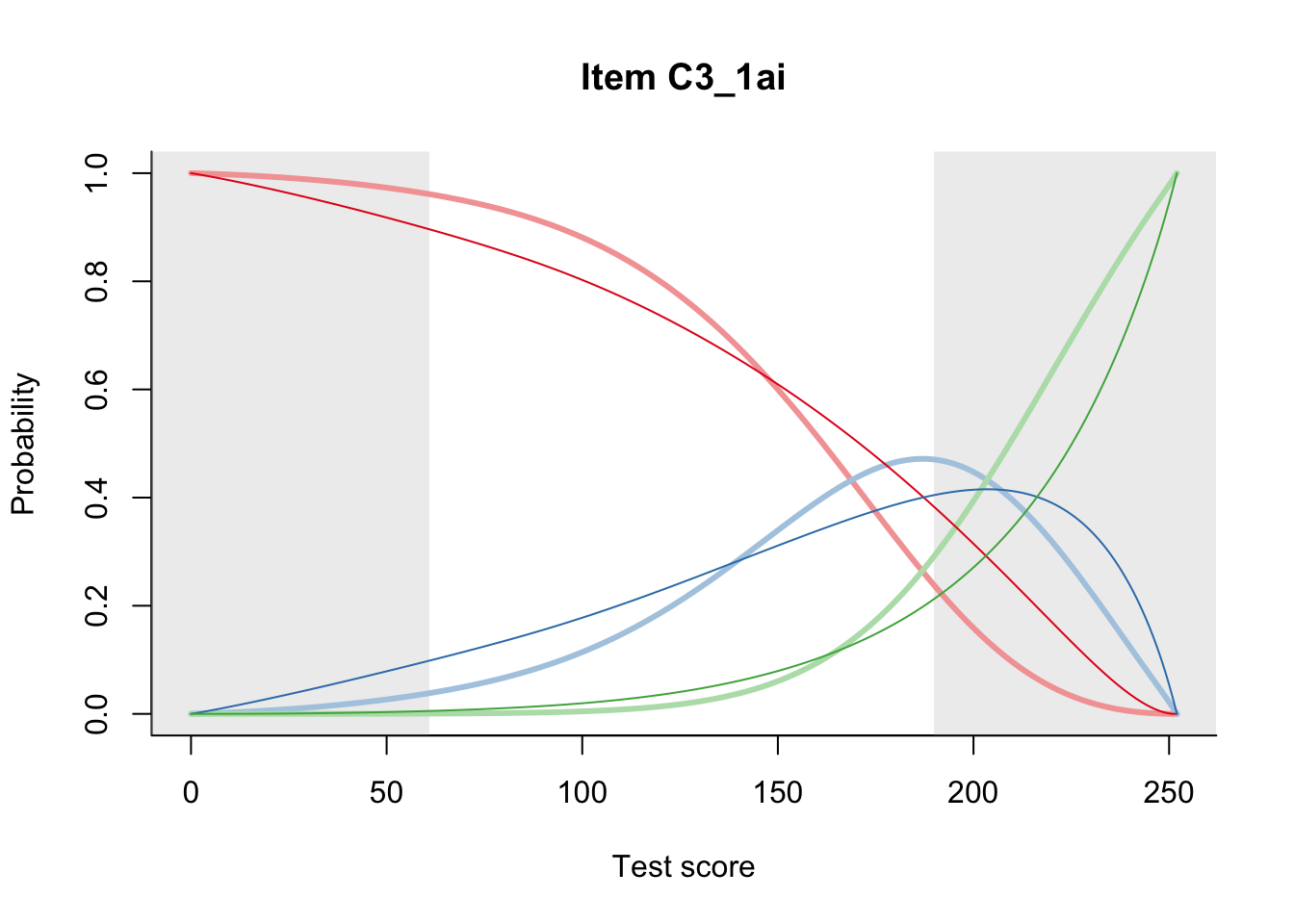
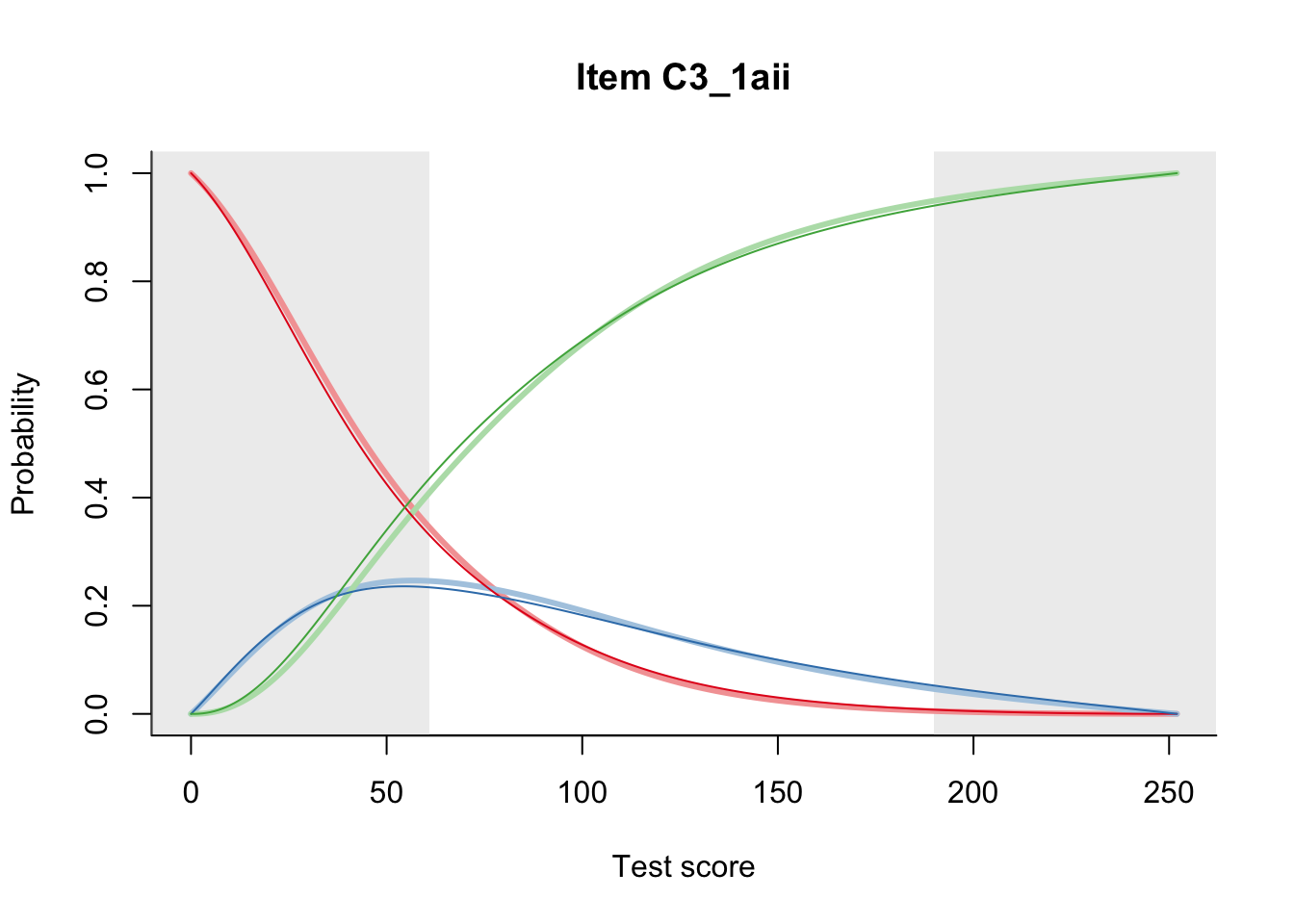
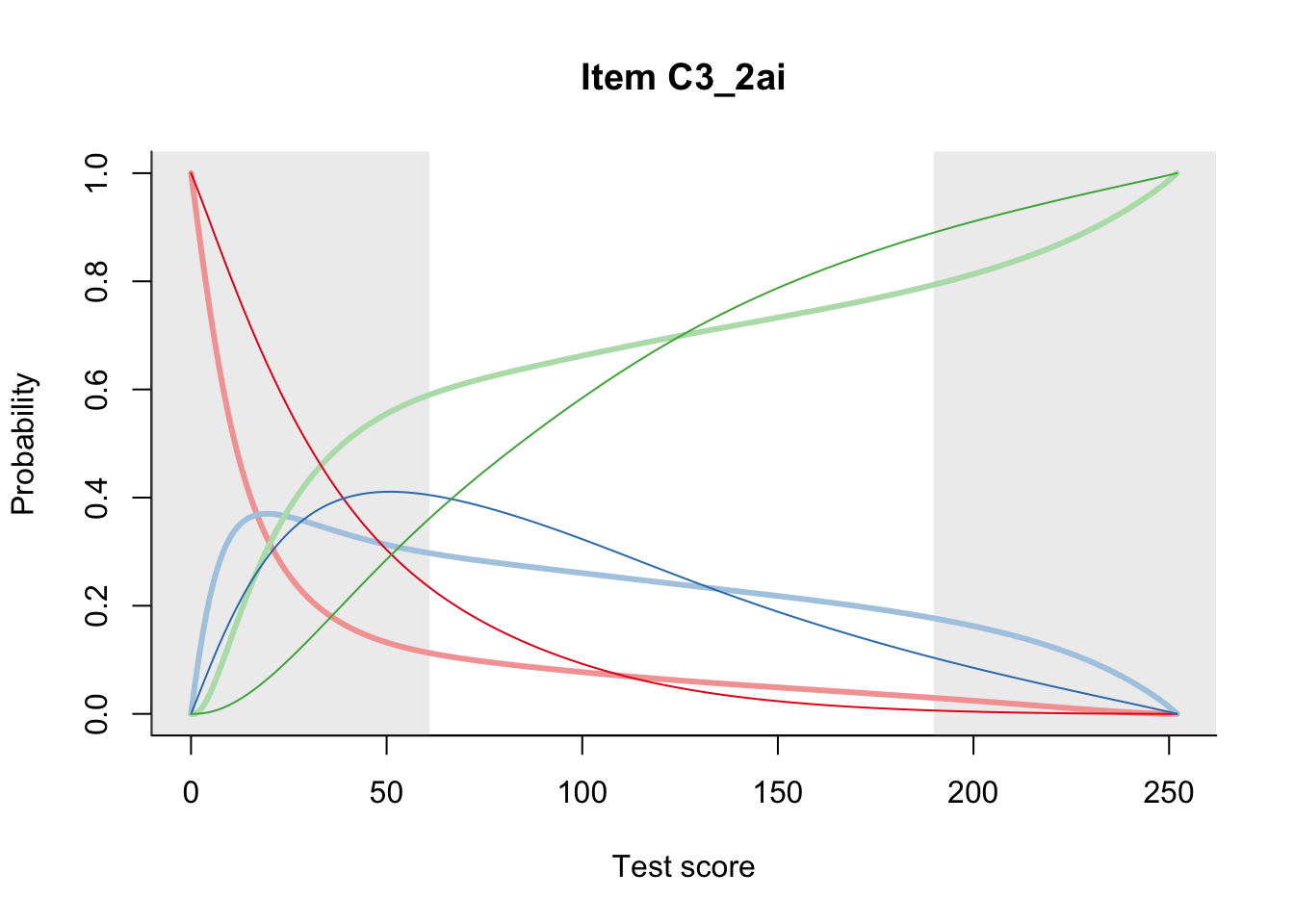
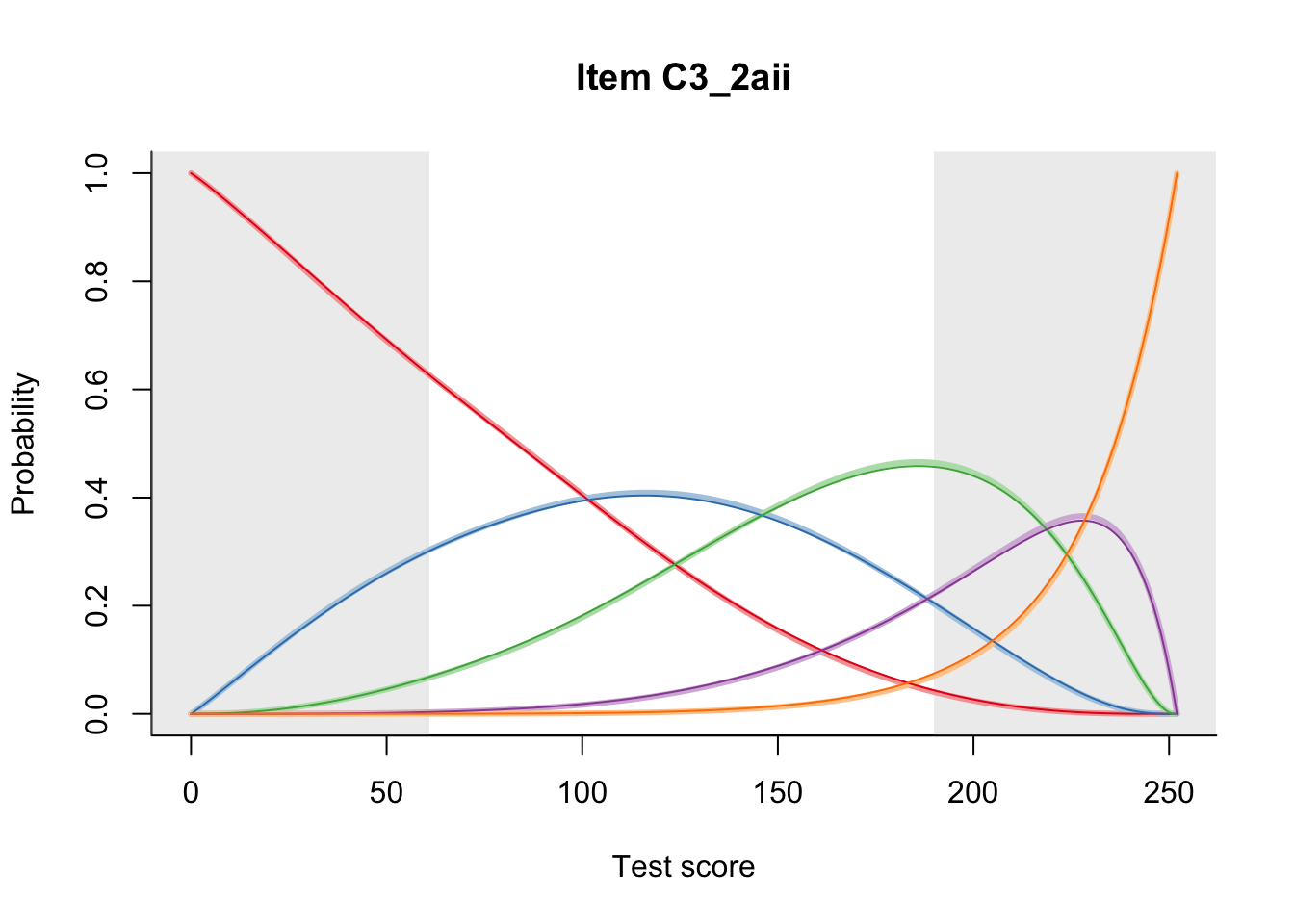
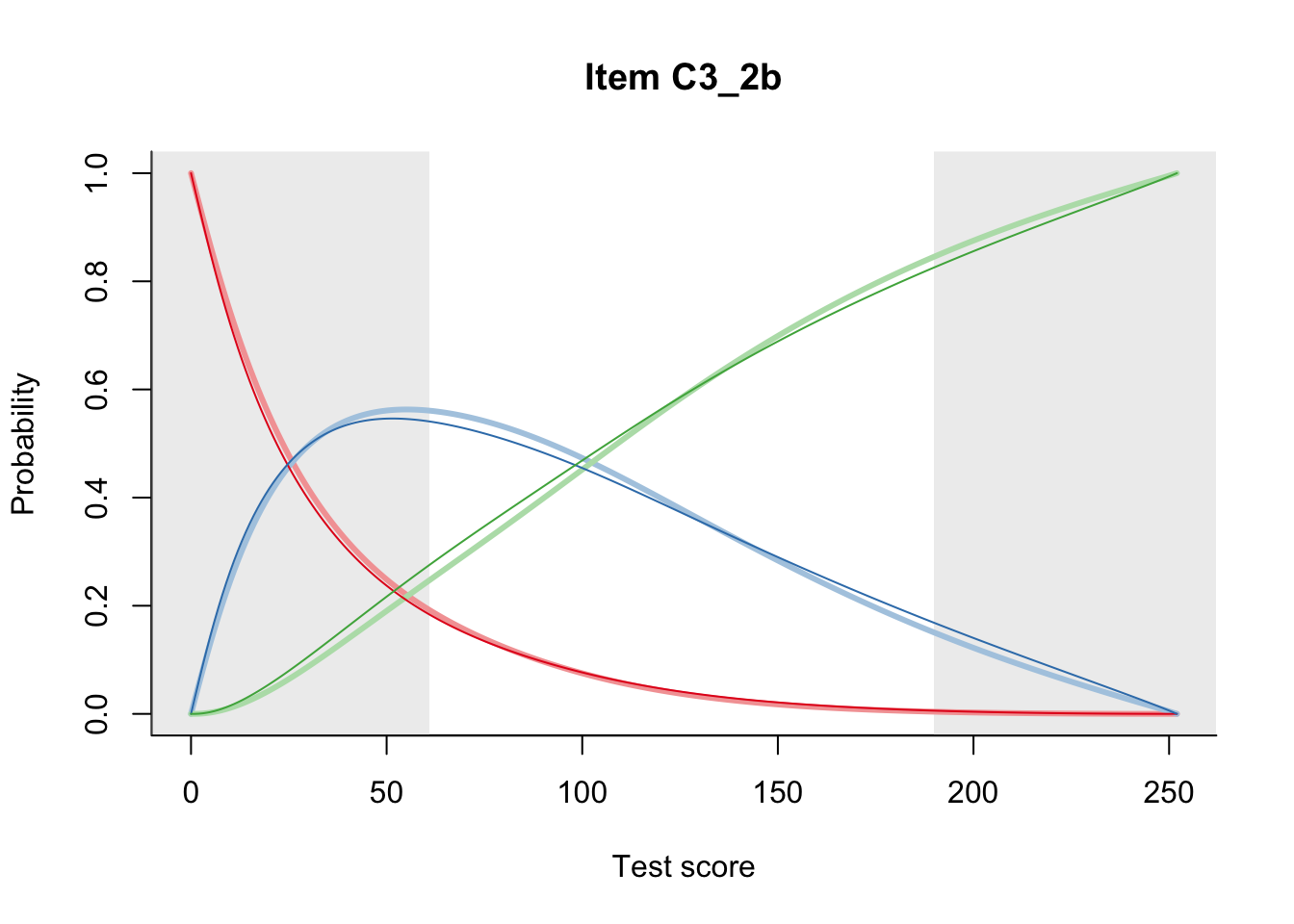
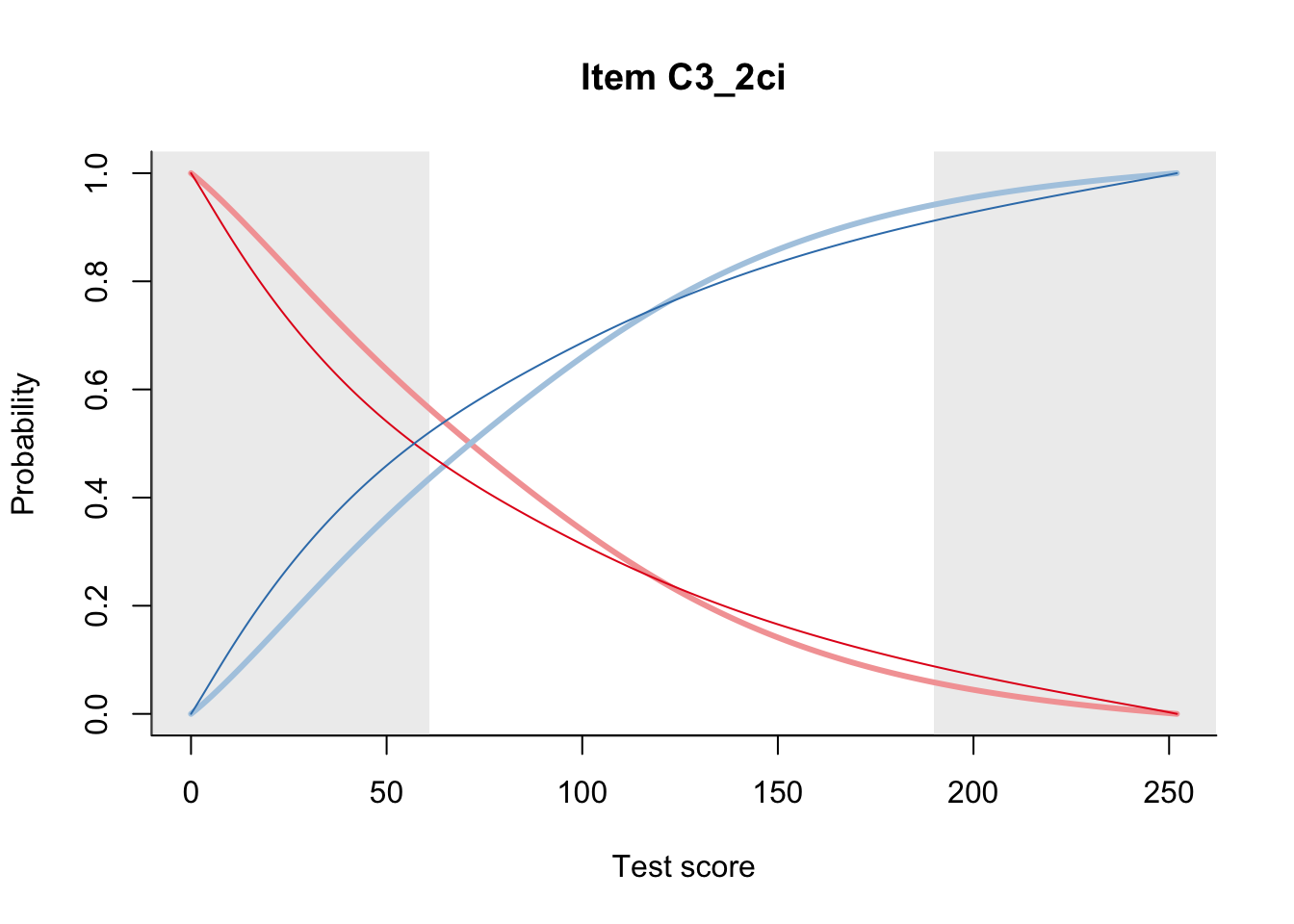
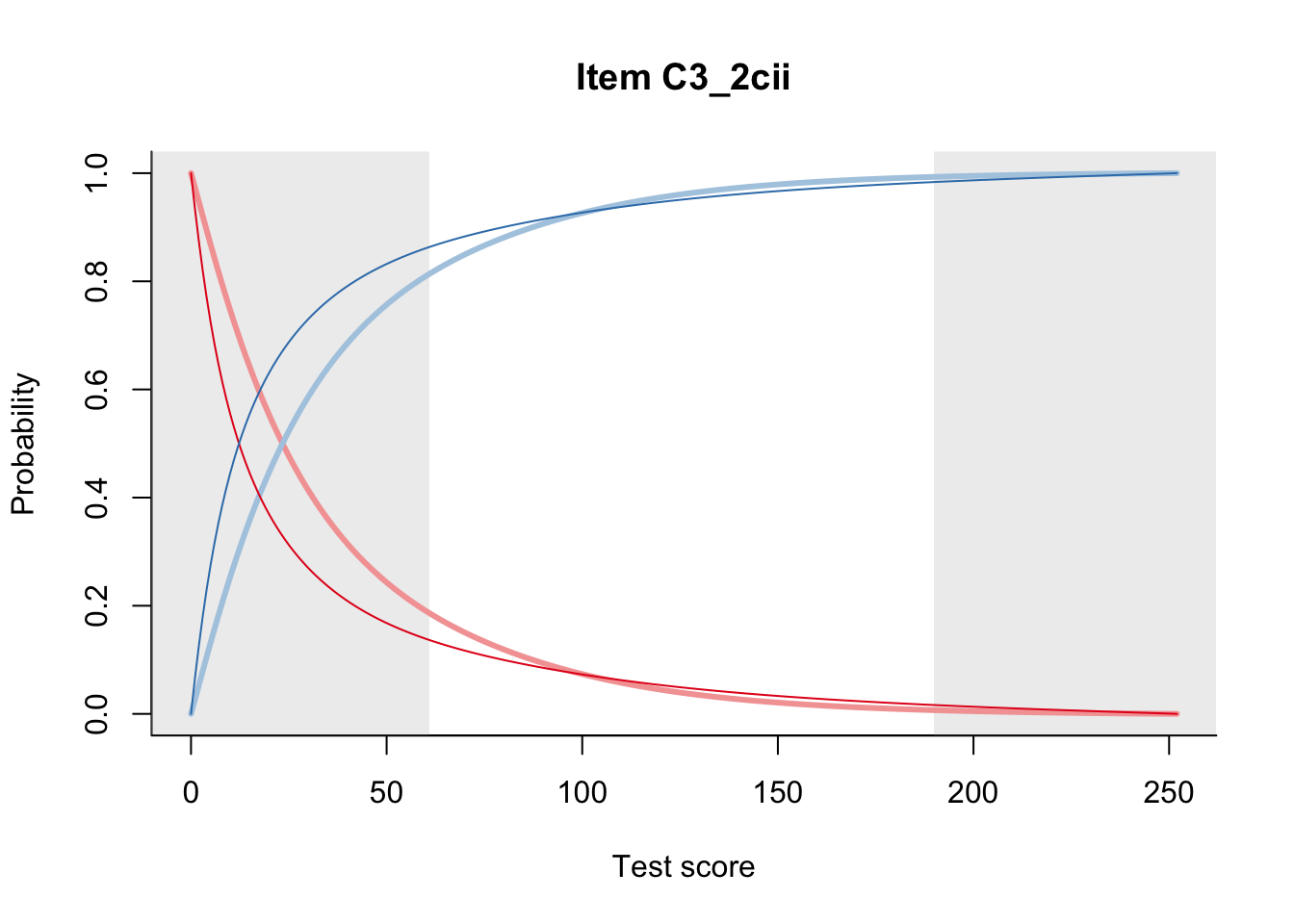
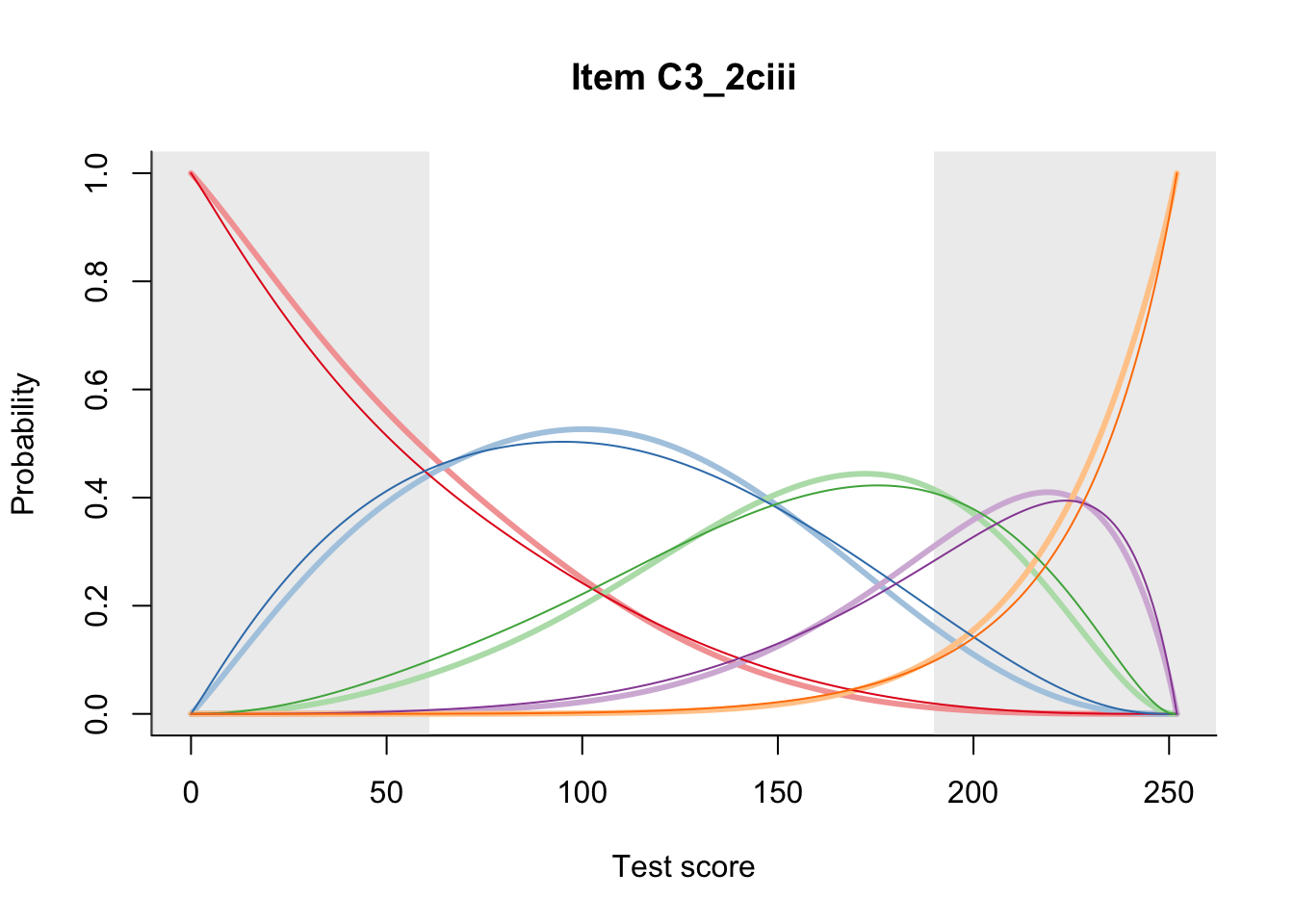
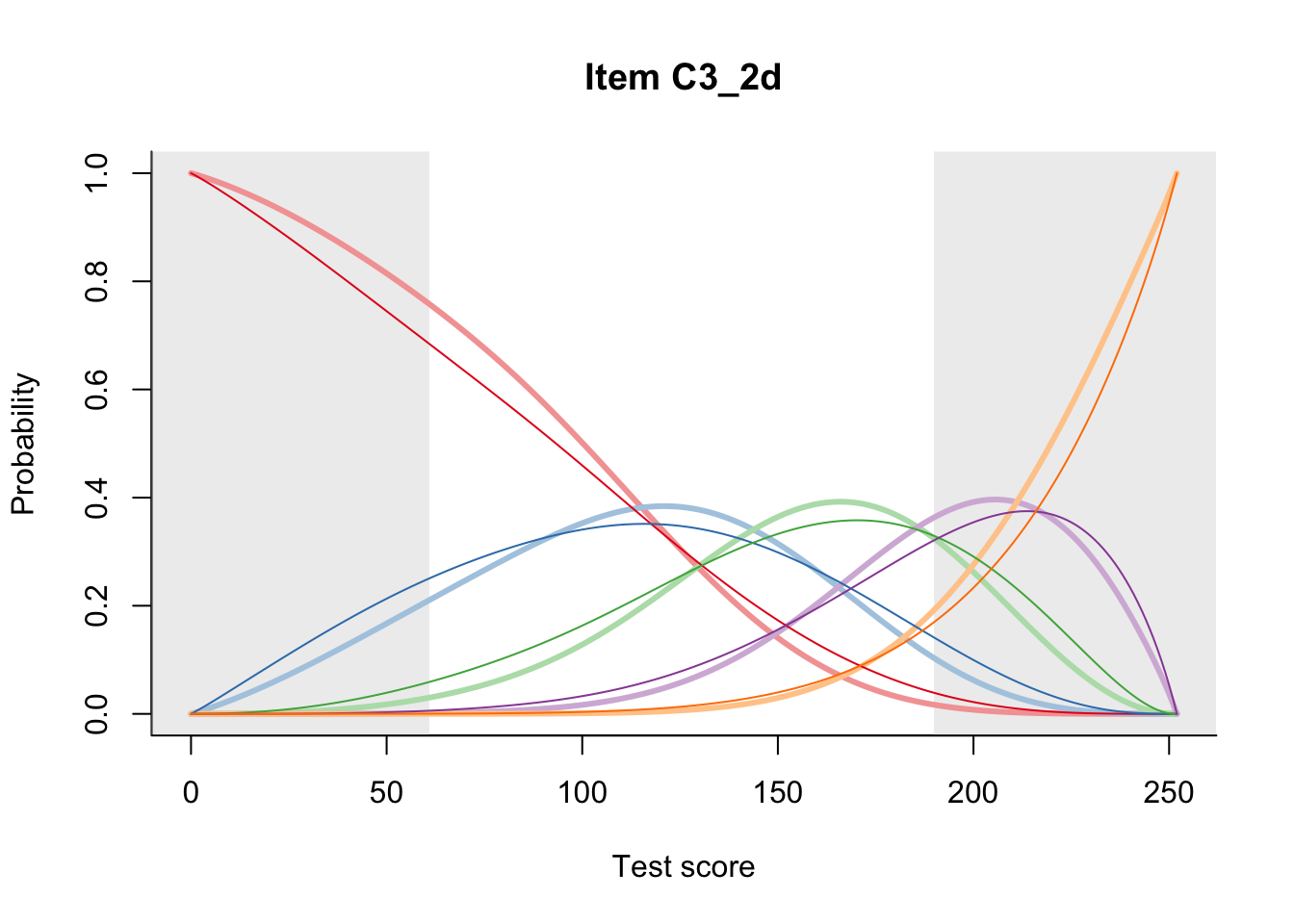
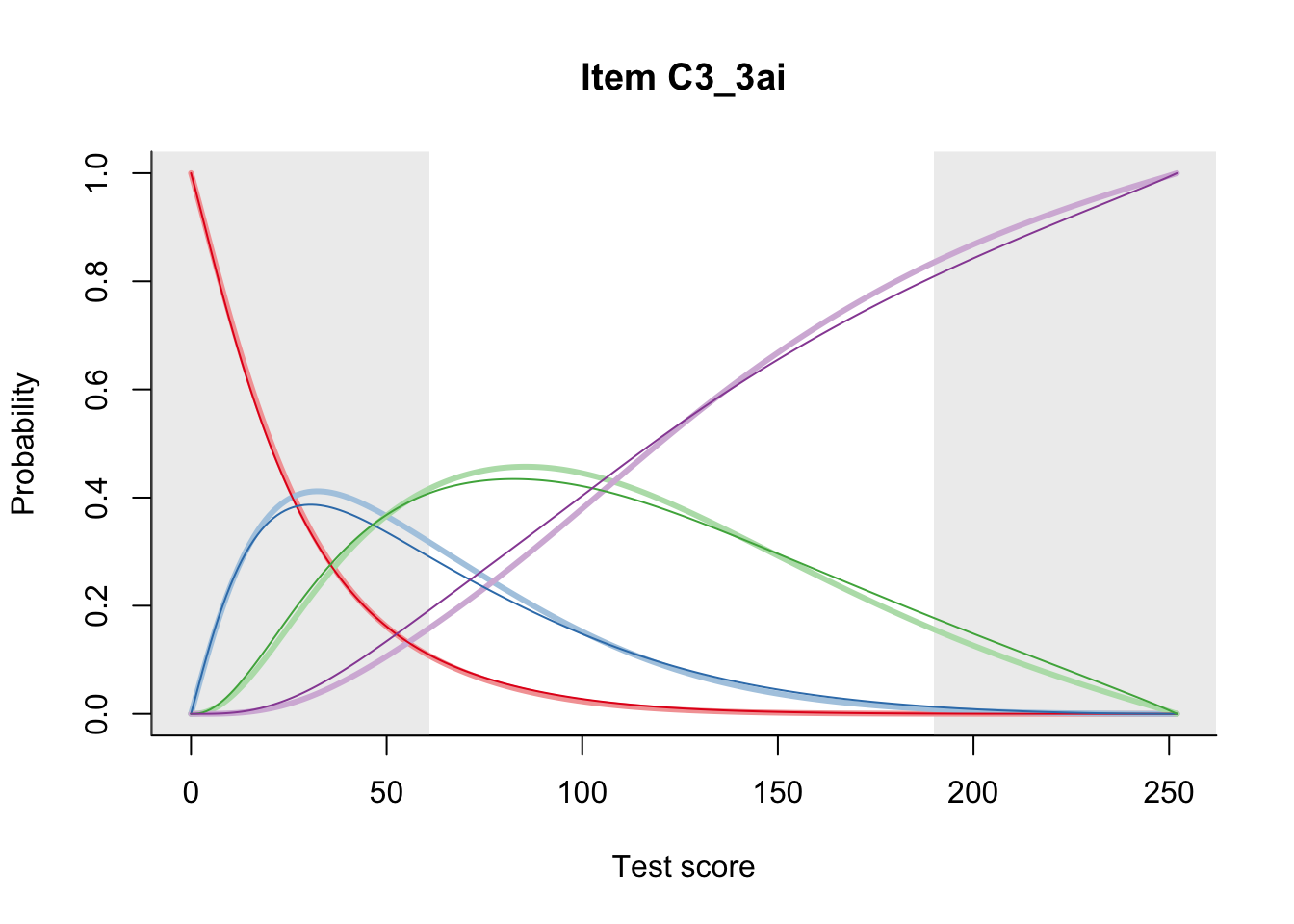
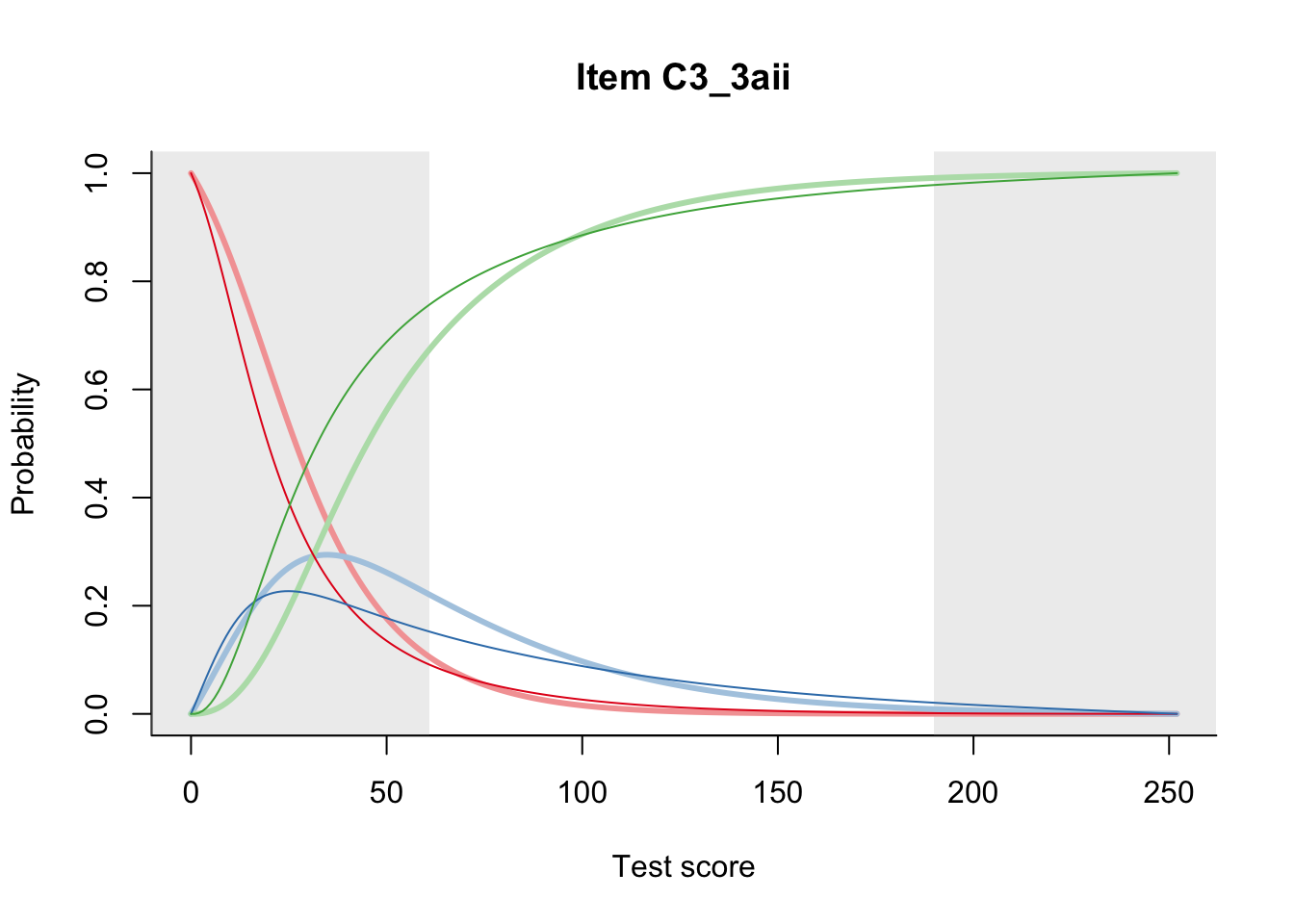
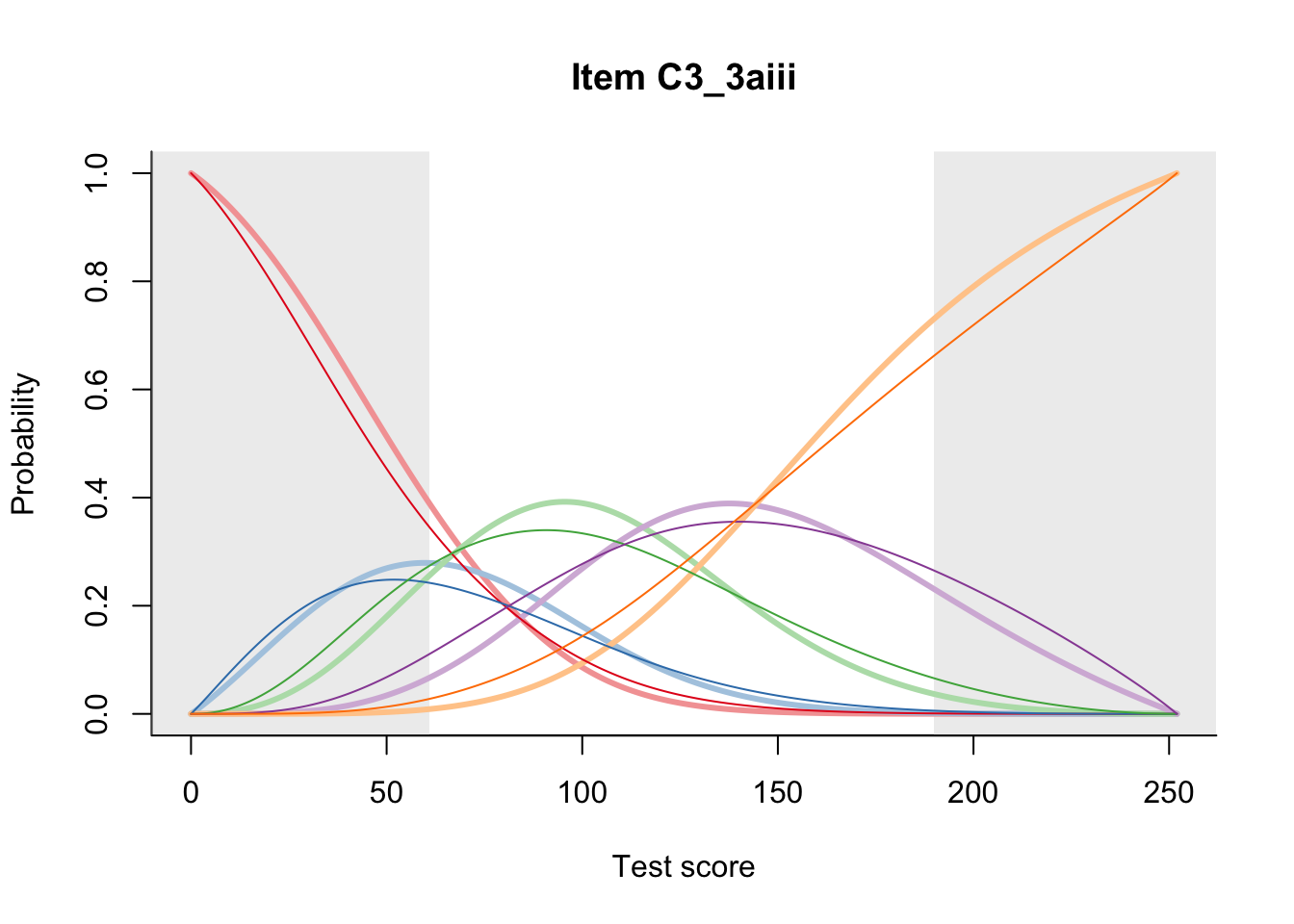
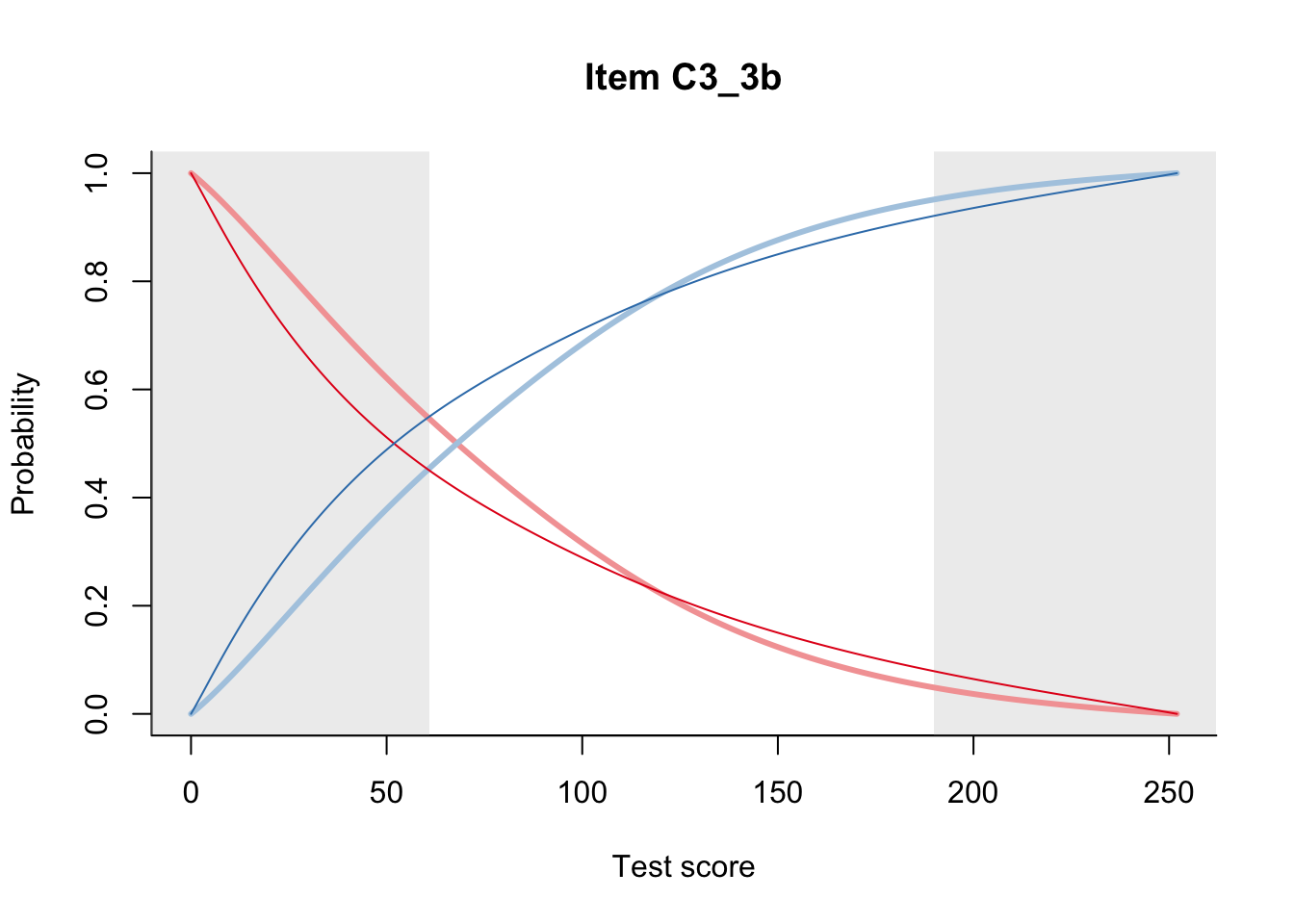
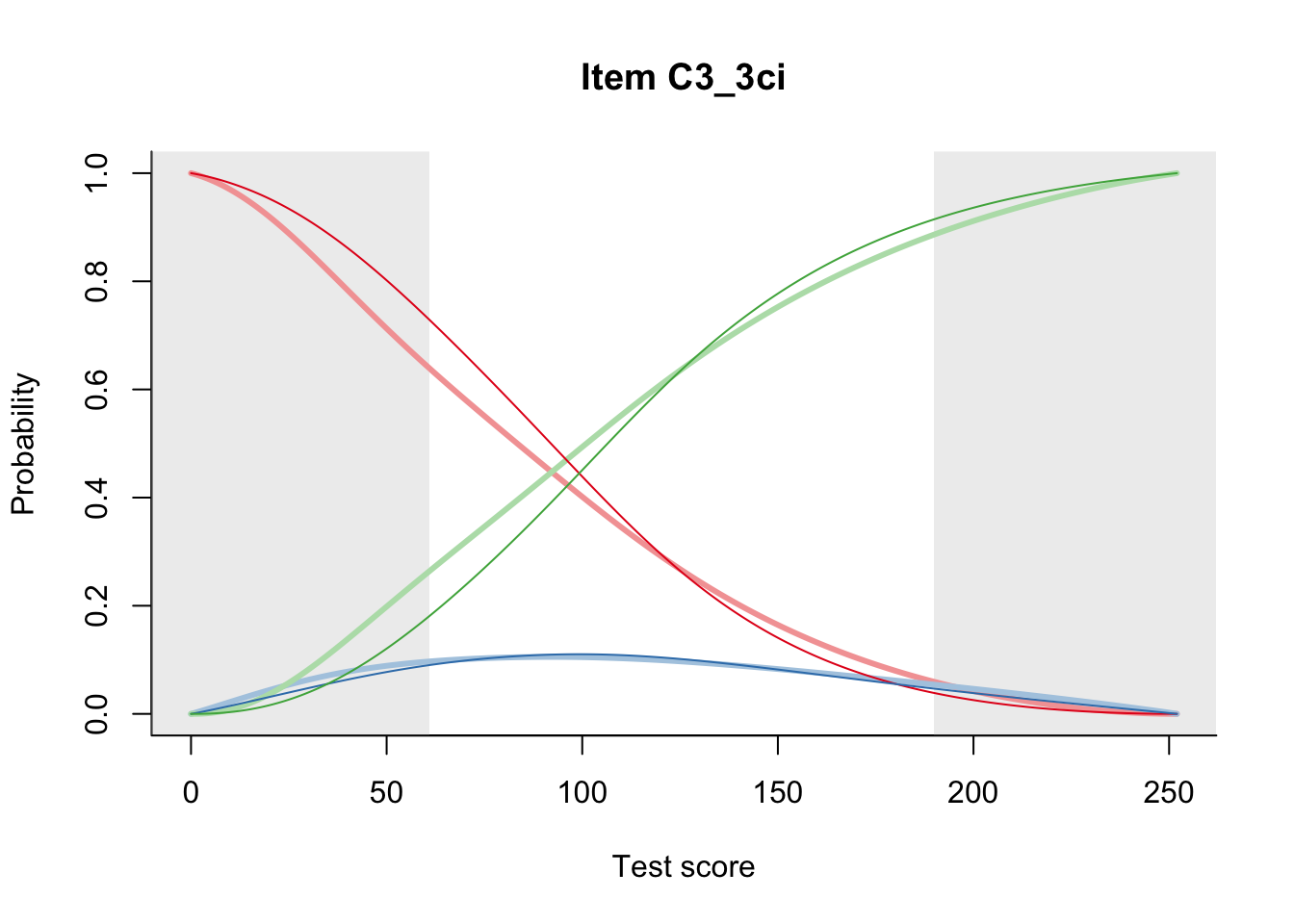
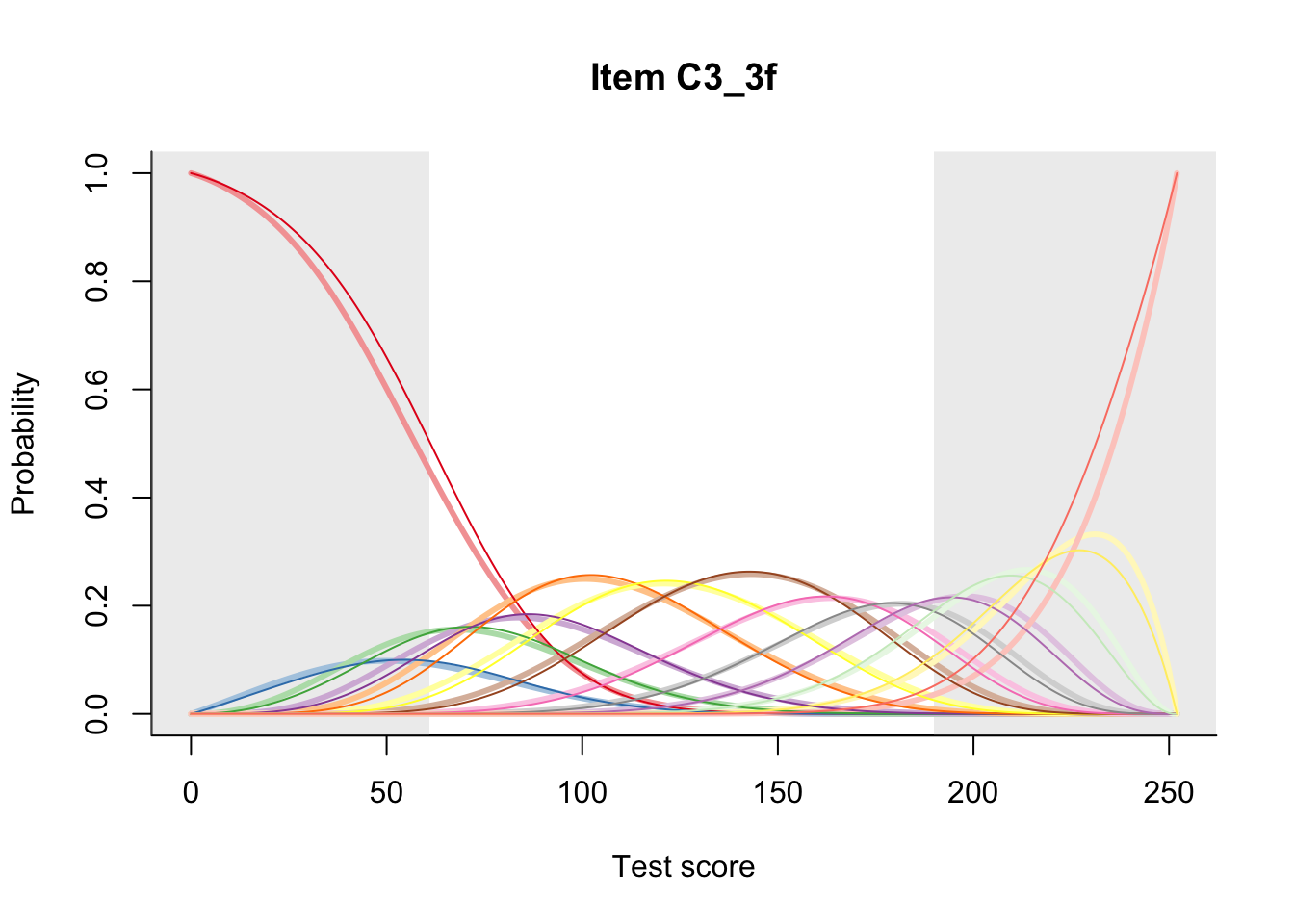
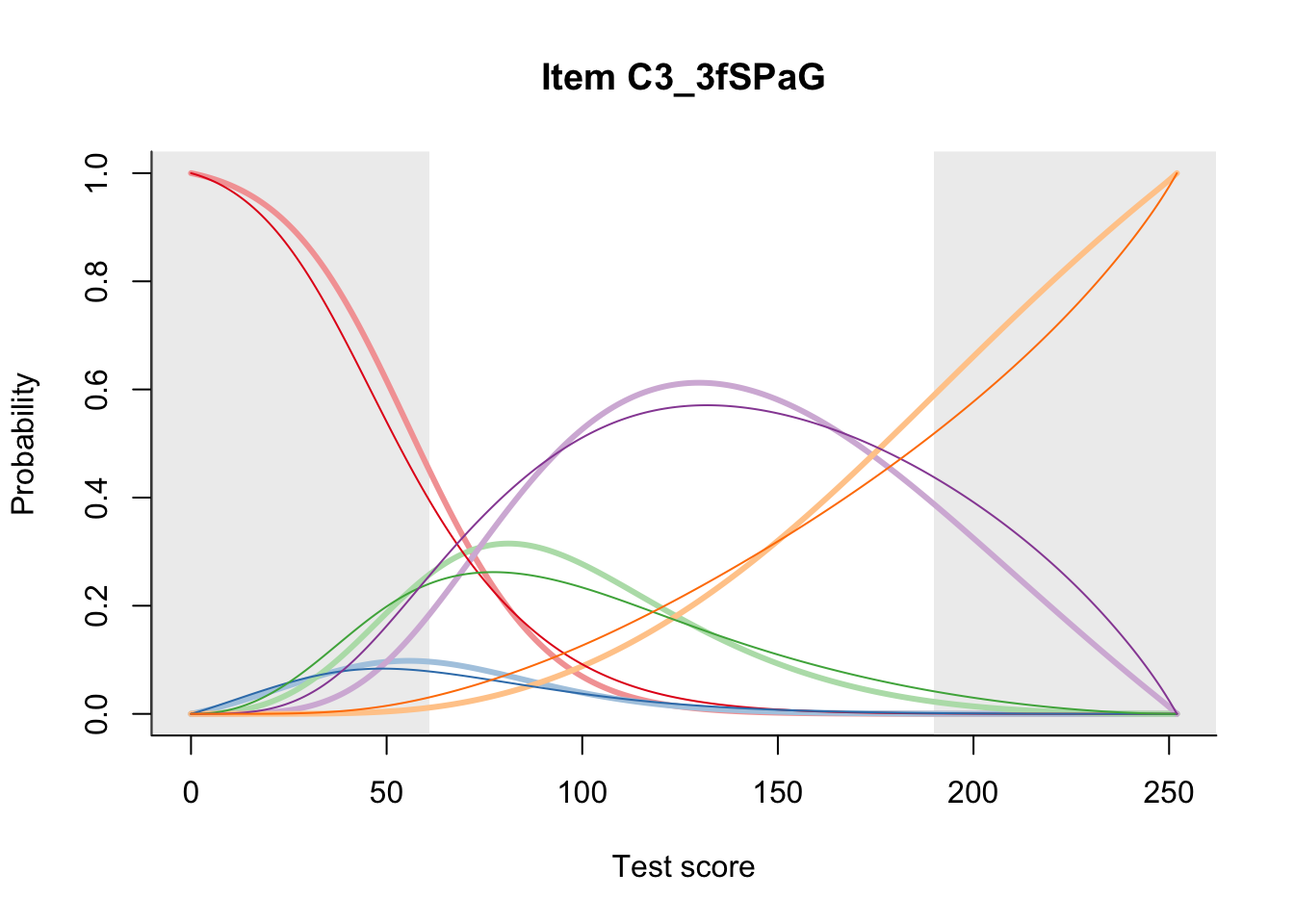
27.2 Item Difficulty
m = fit_inter(db)
n_items <- nrow(data_summary)
for(i in 1:n_items){
plot(m, data_summary$Variable[i], summate=FALSE, show.observed=FALSE)
}
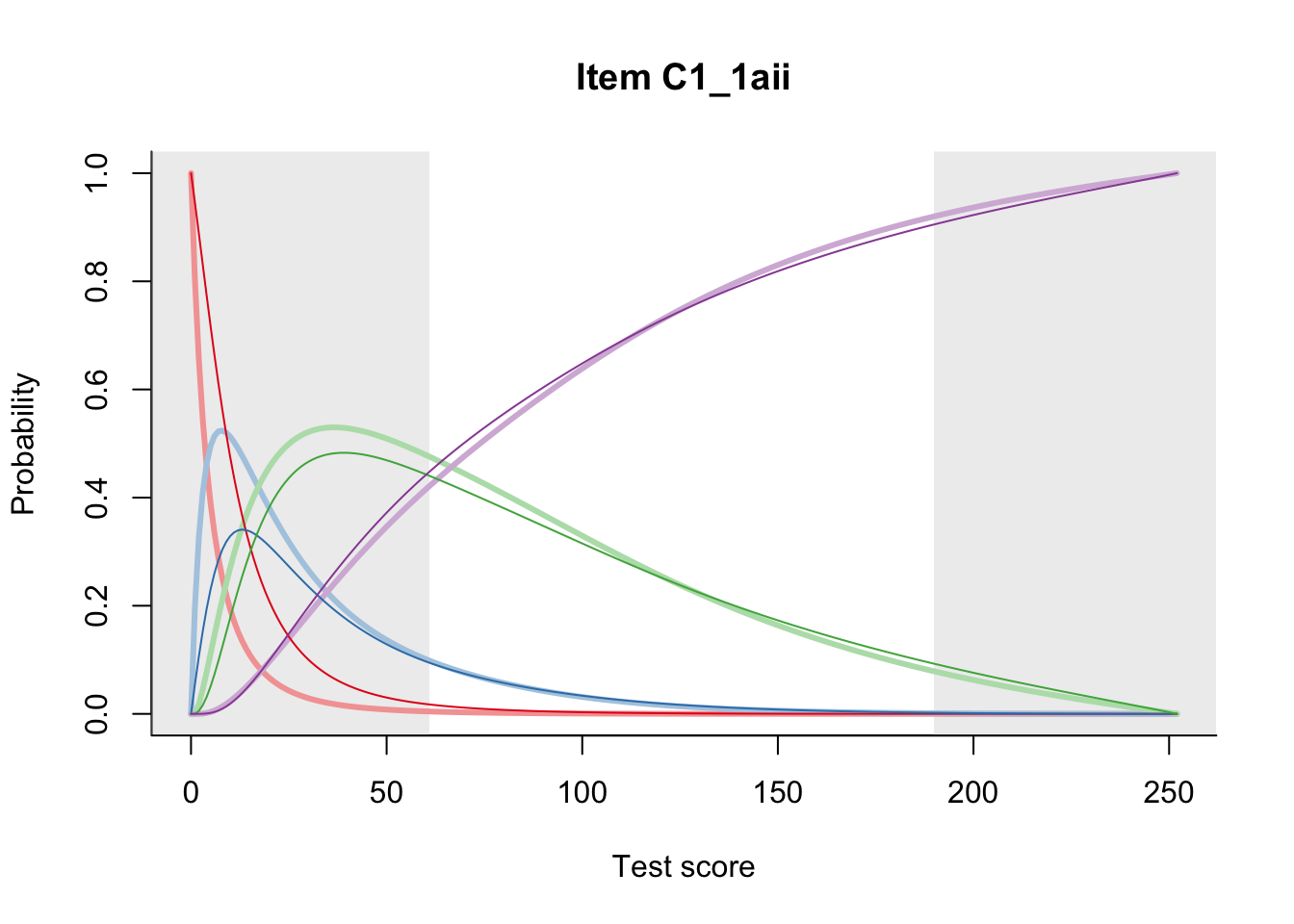
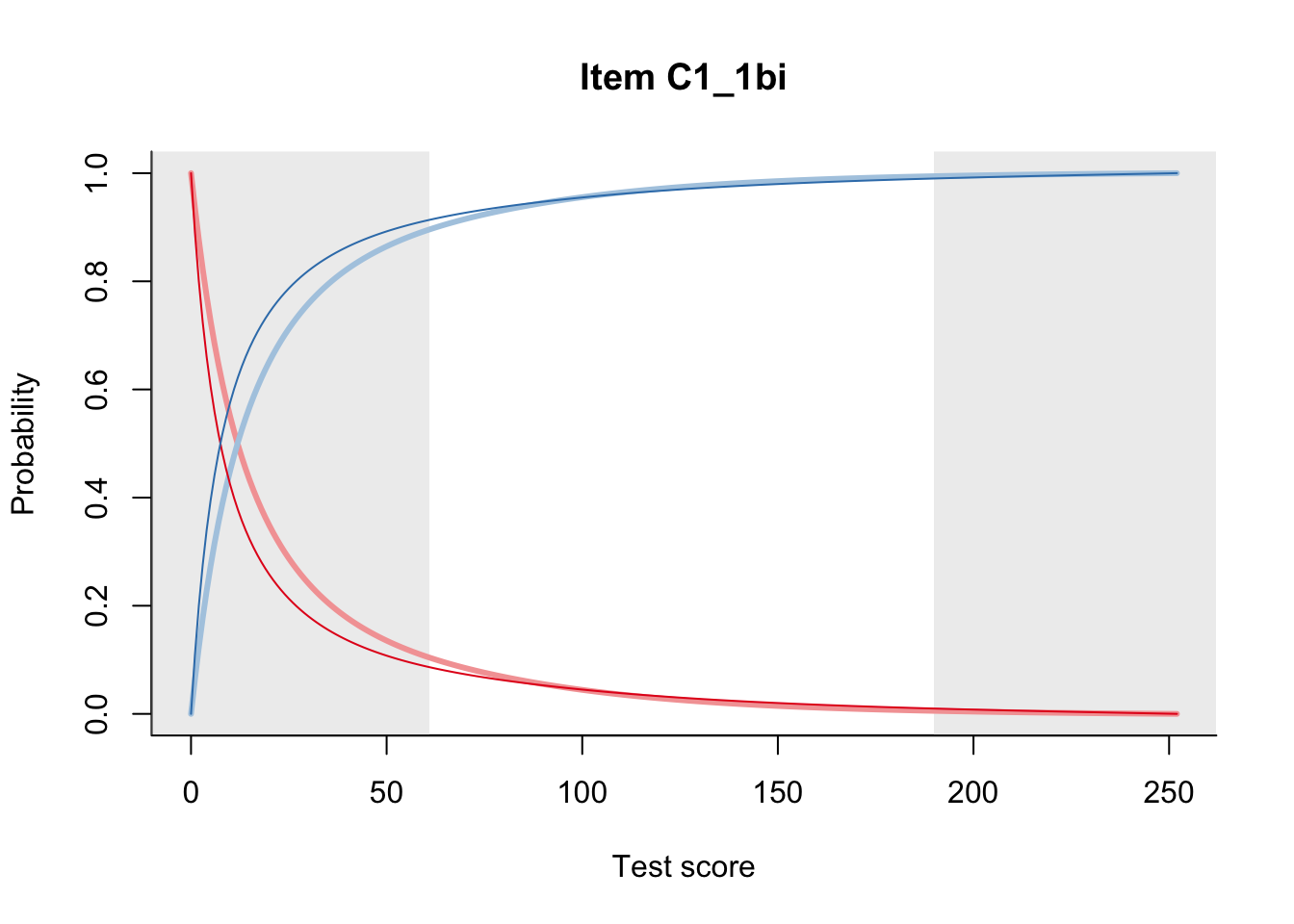
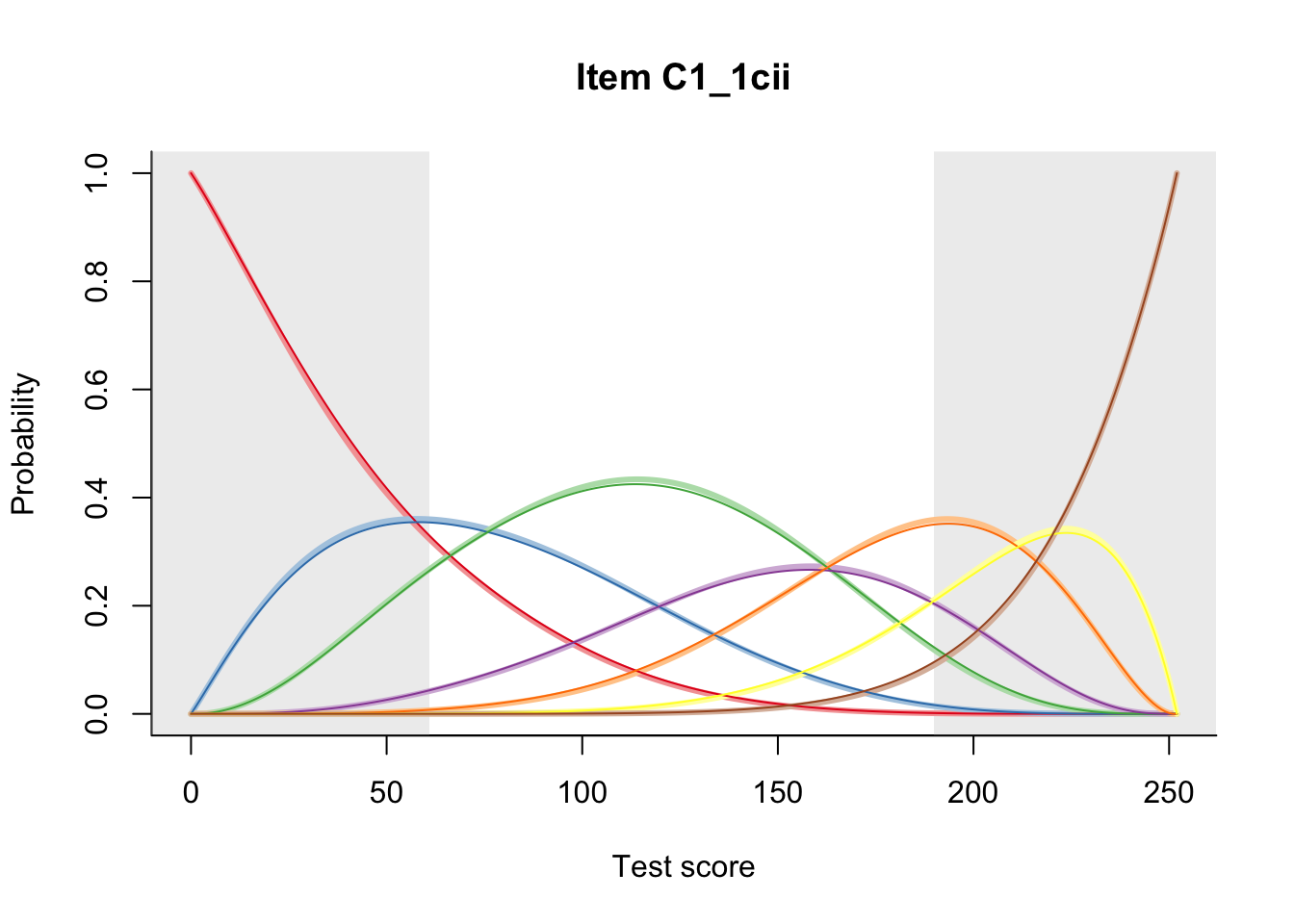
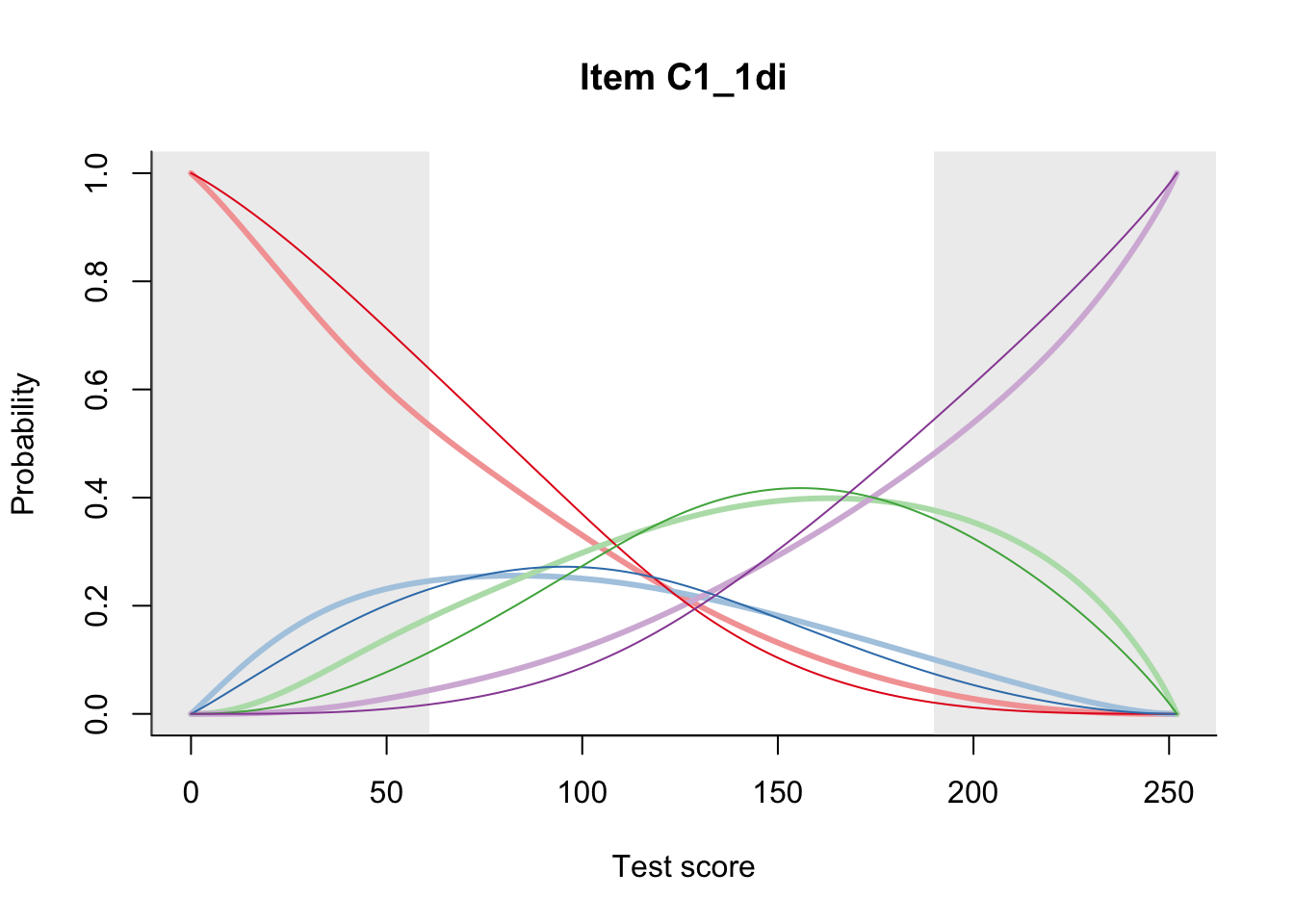
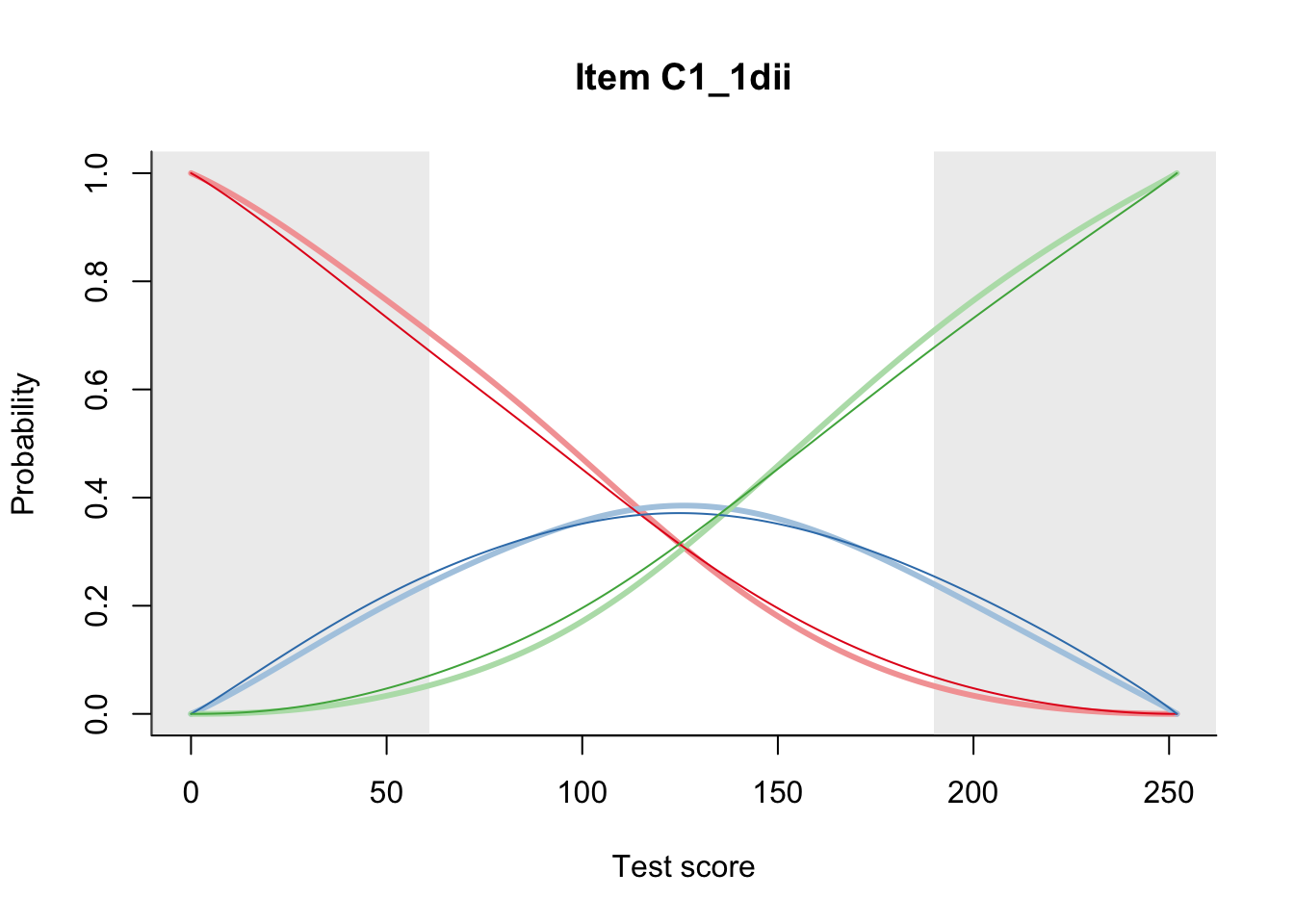
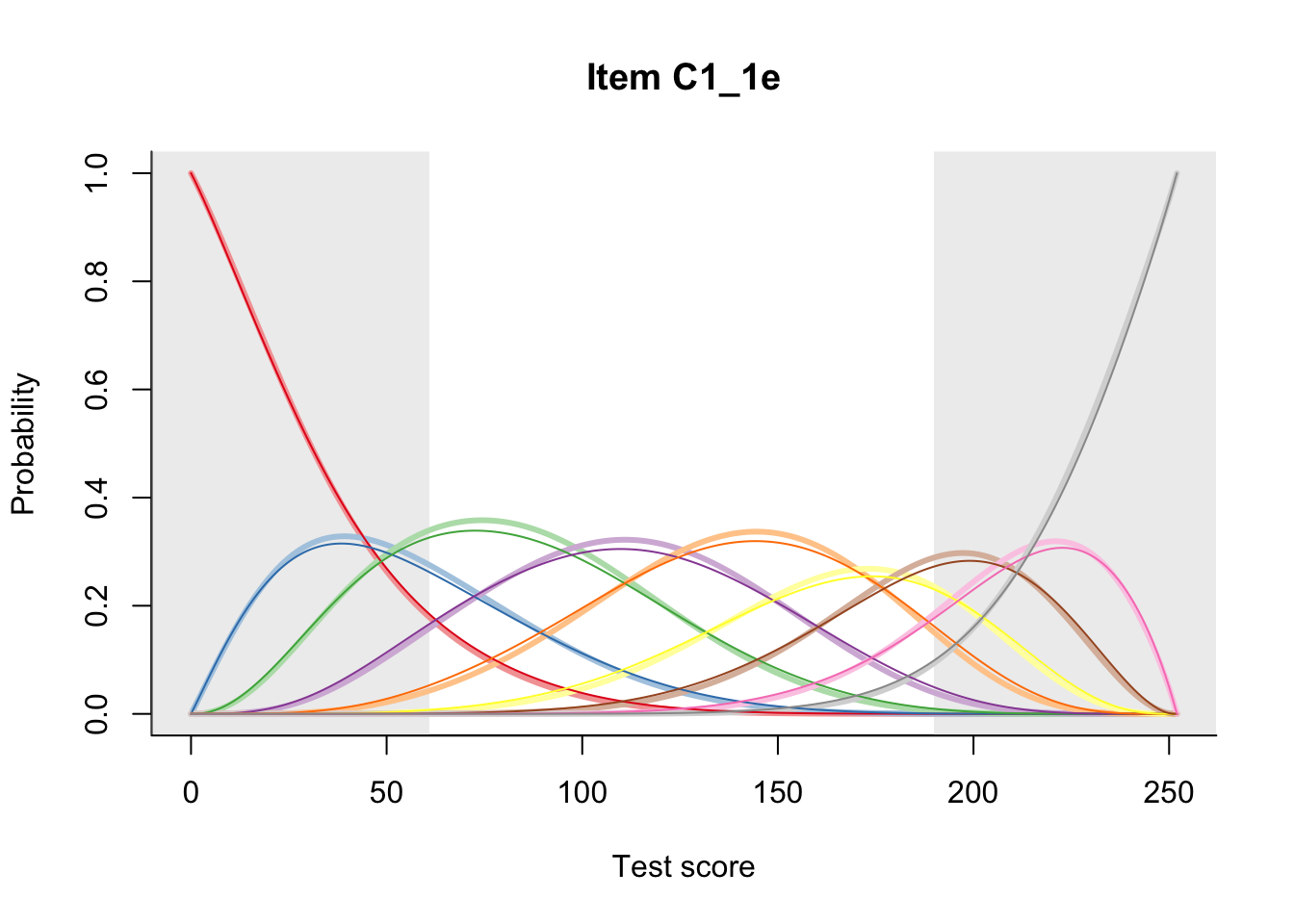
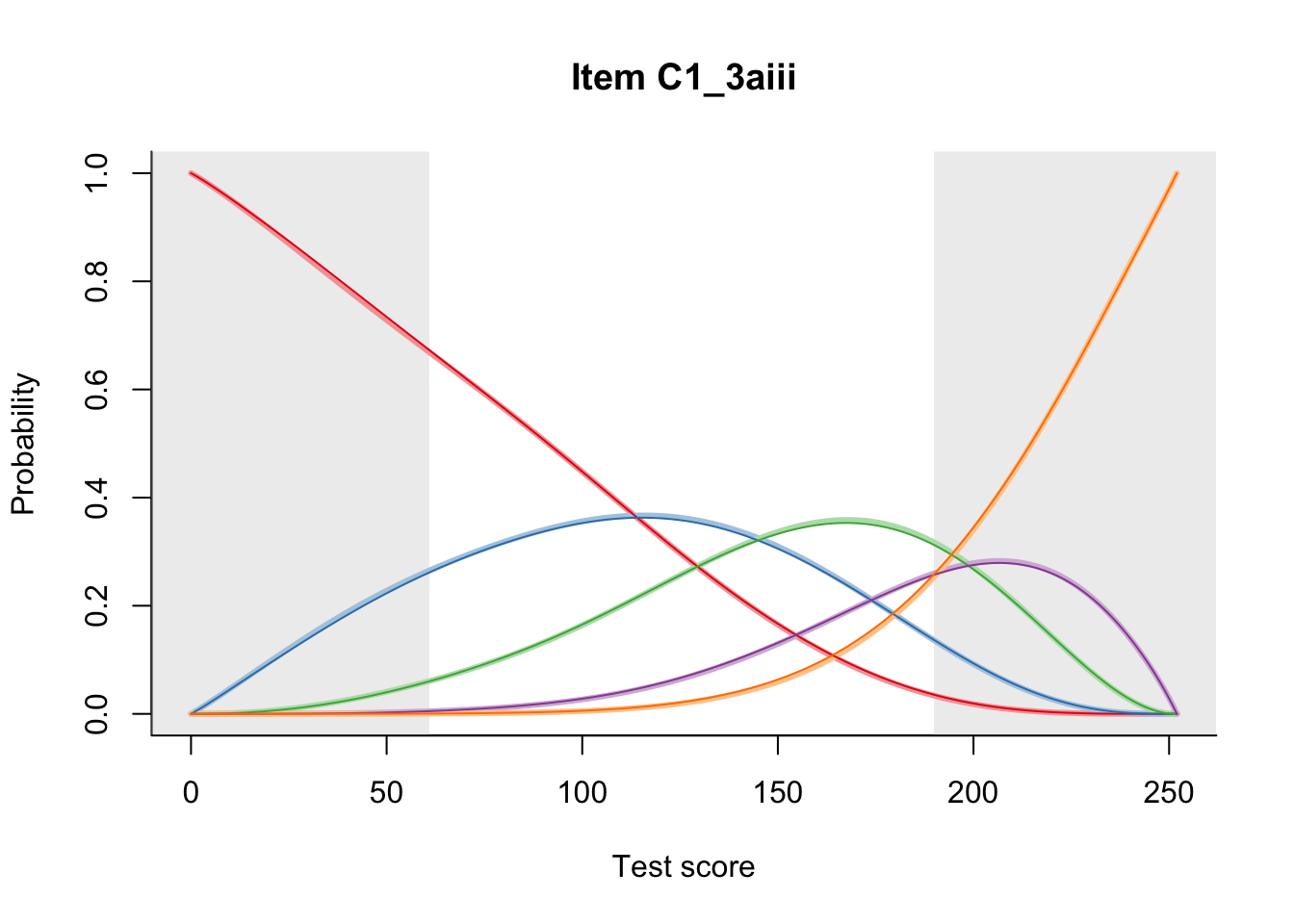
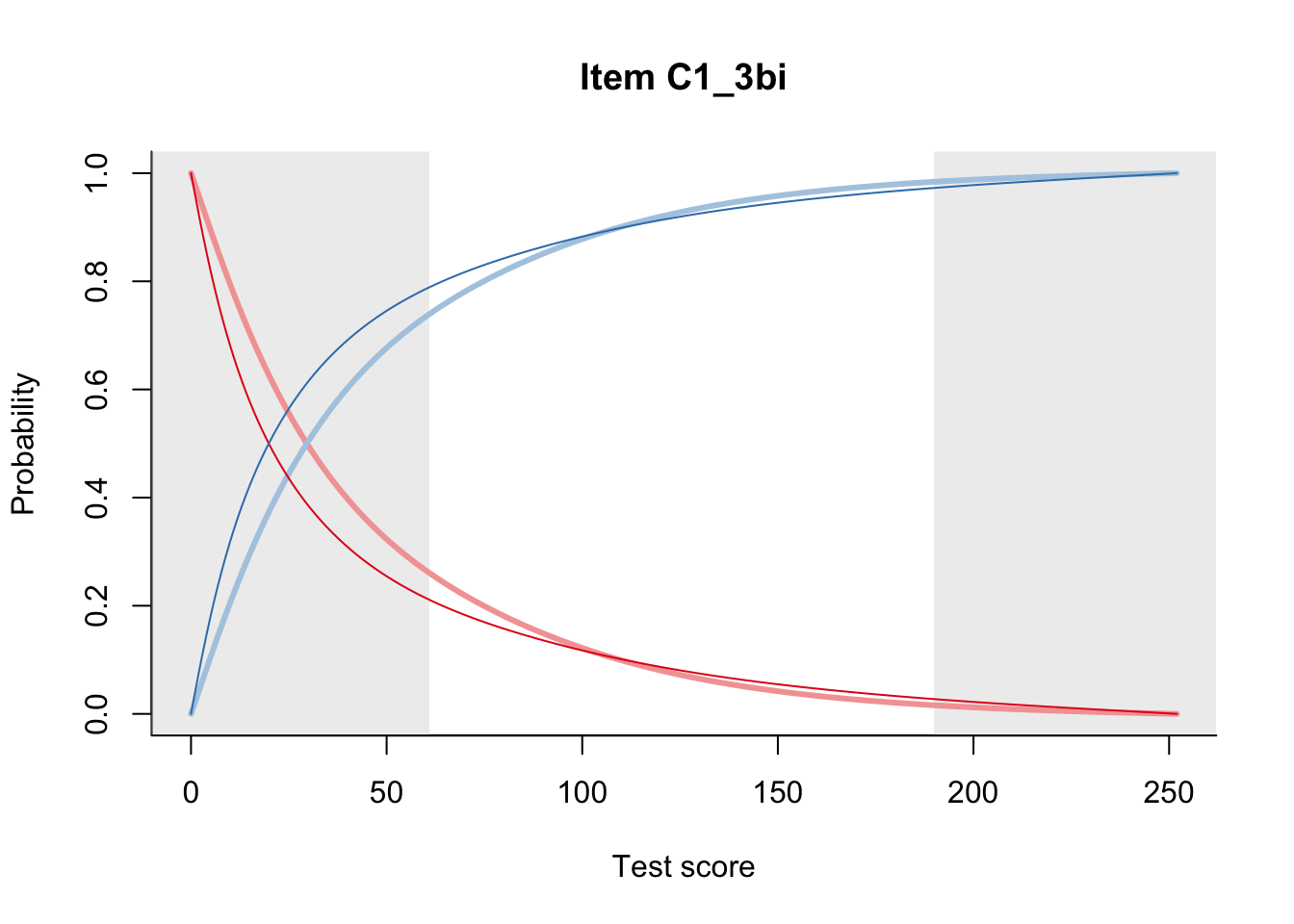
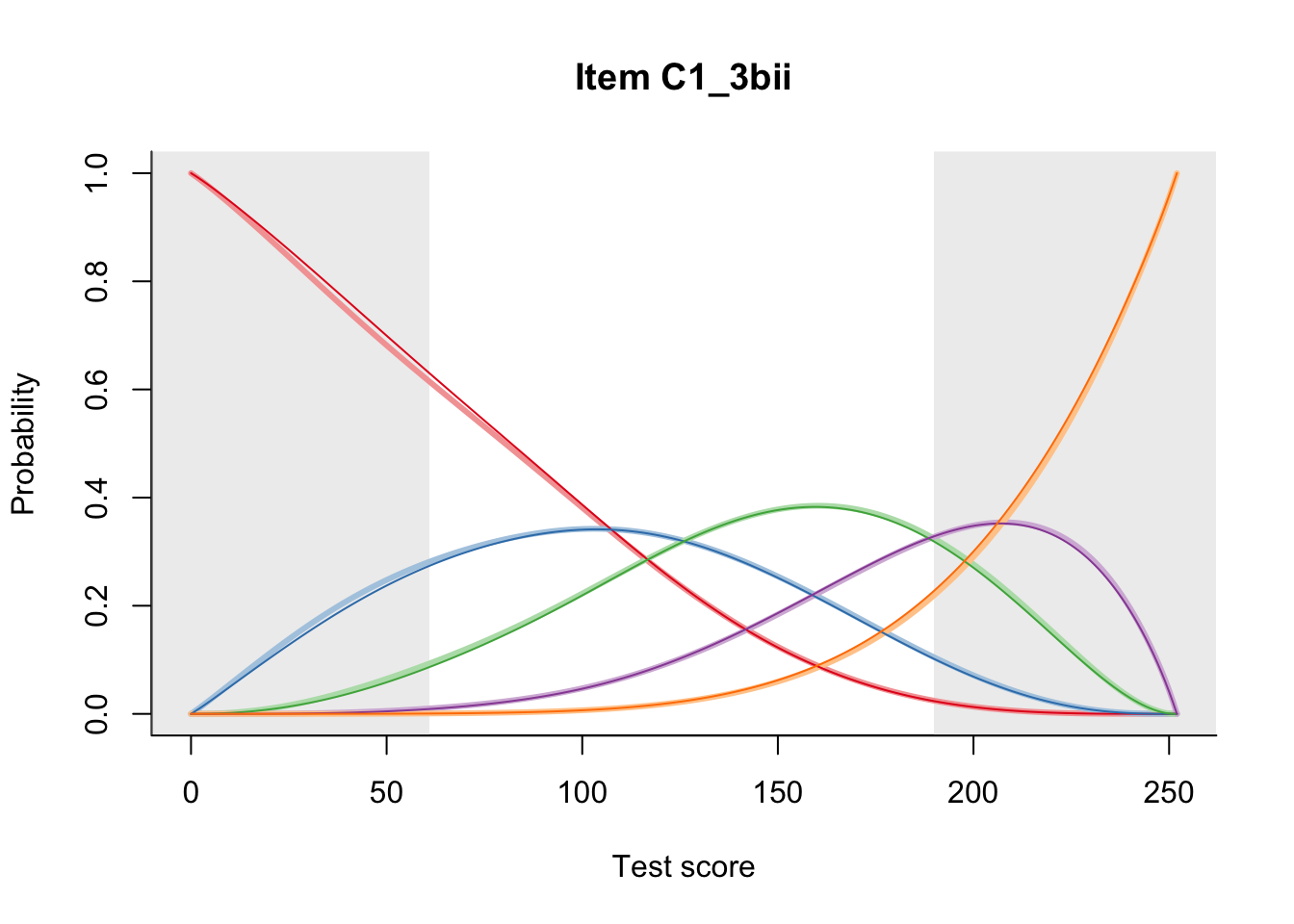
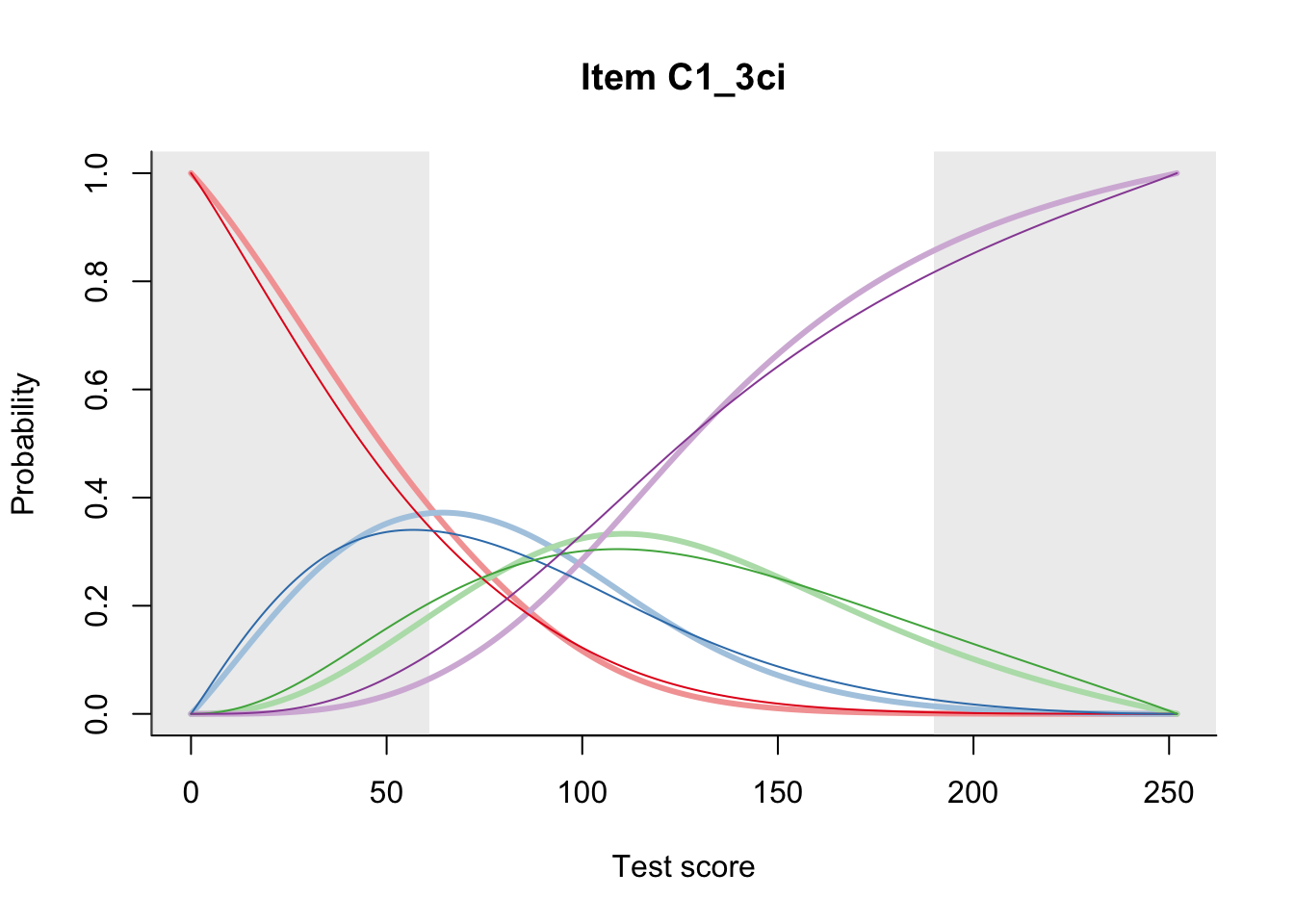
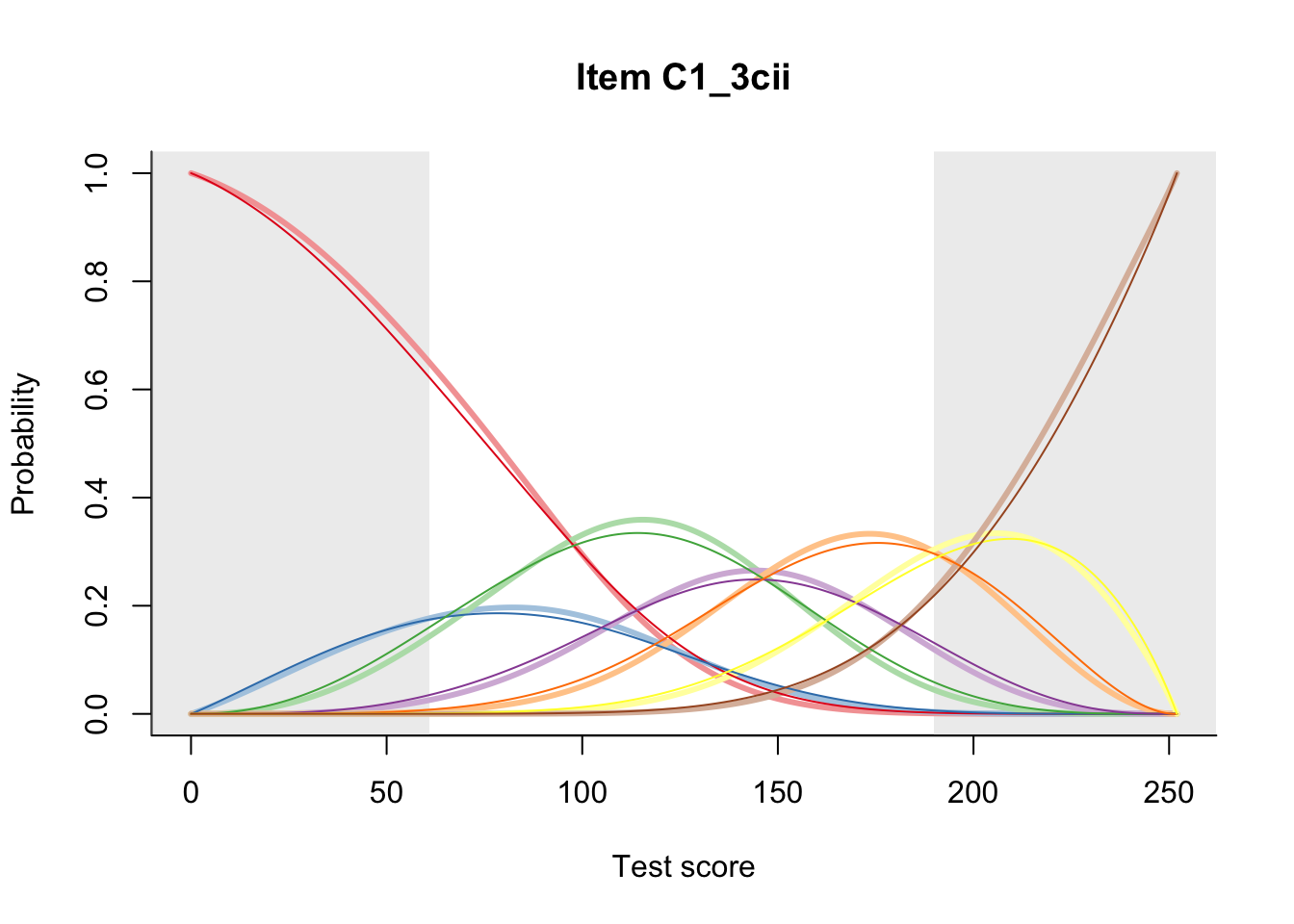
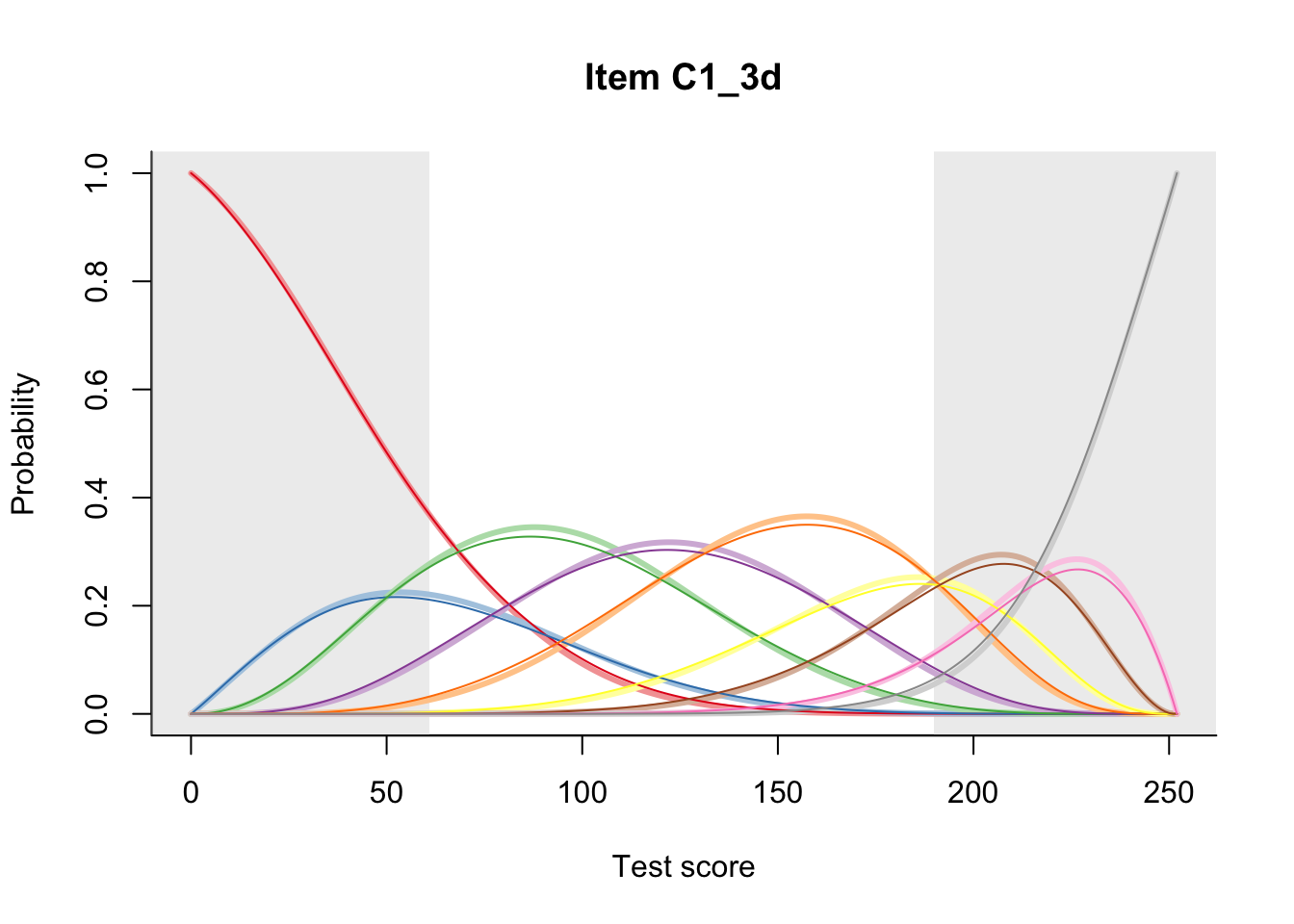
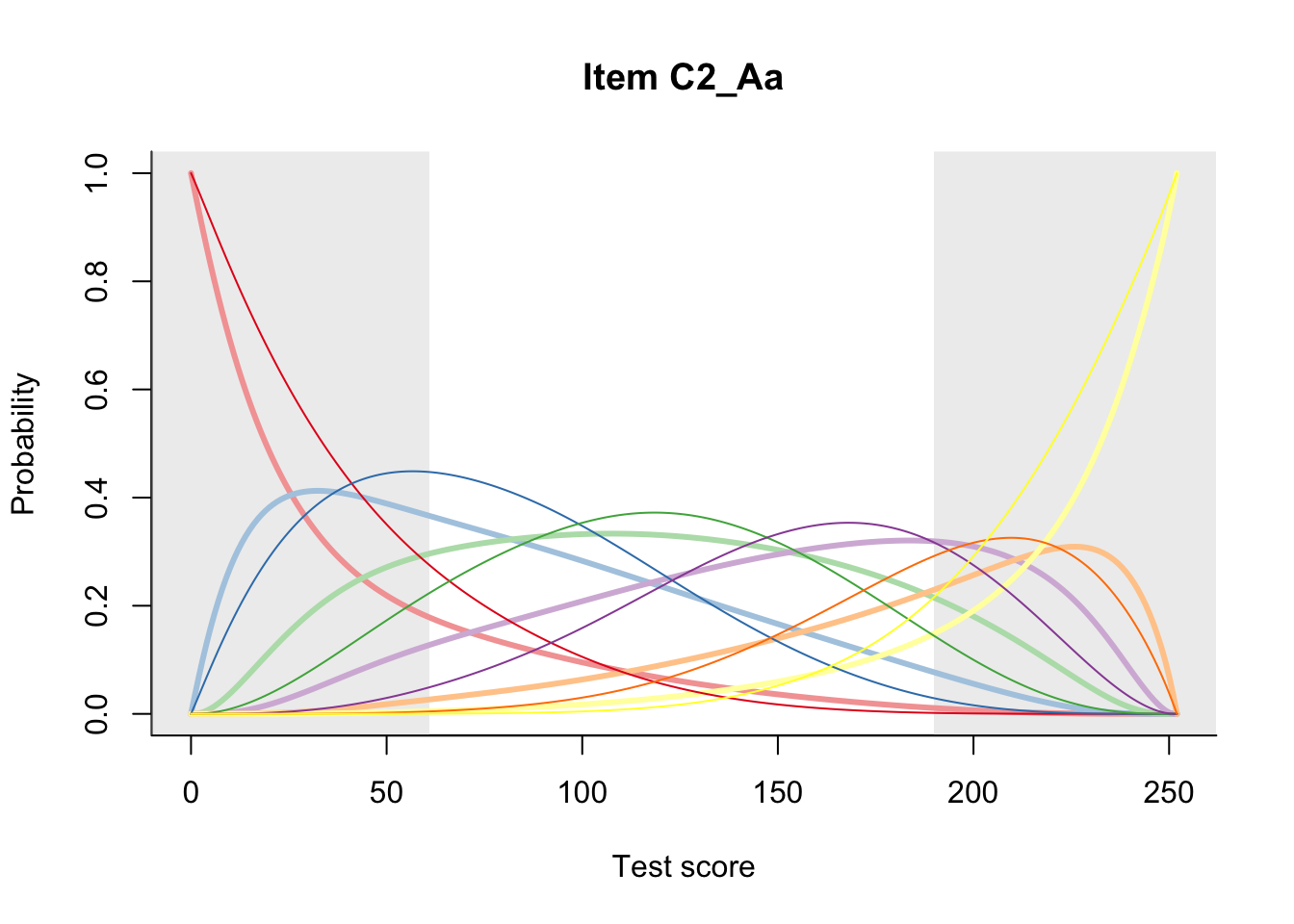
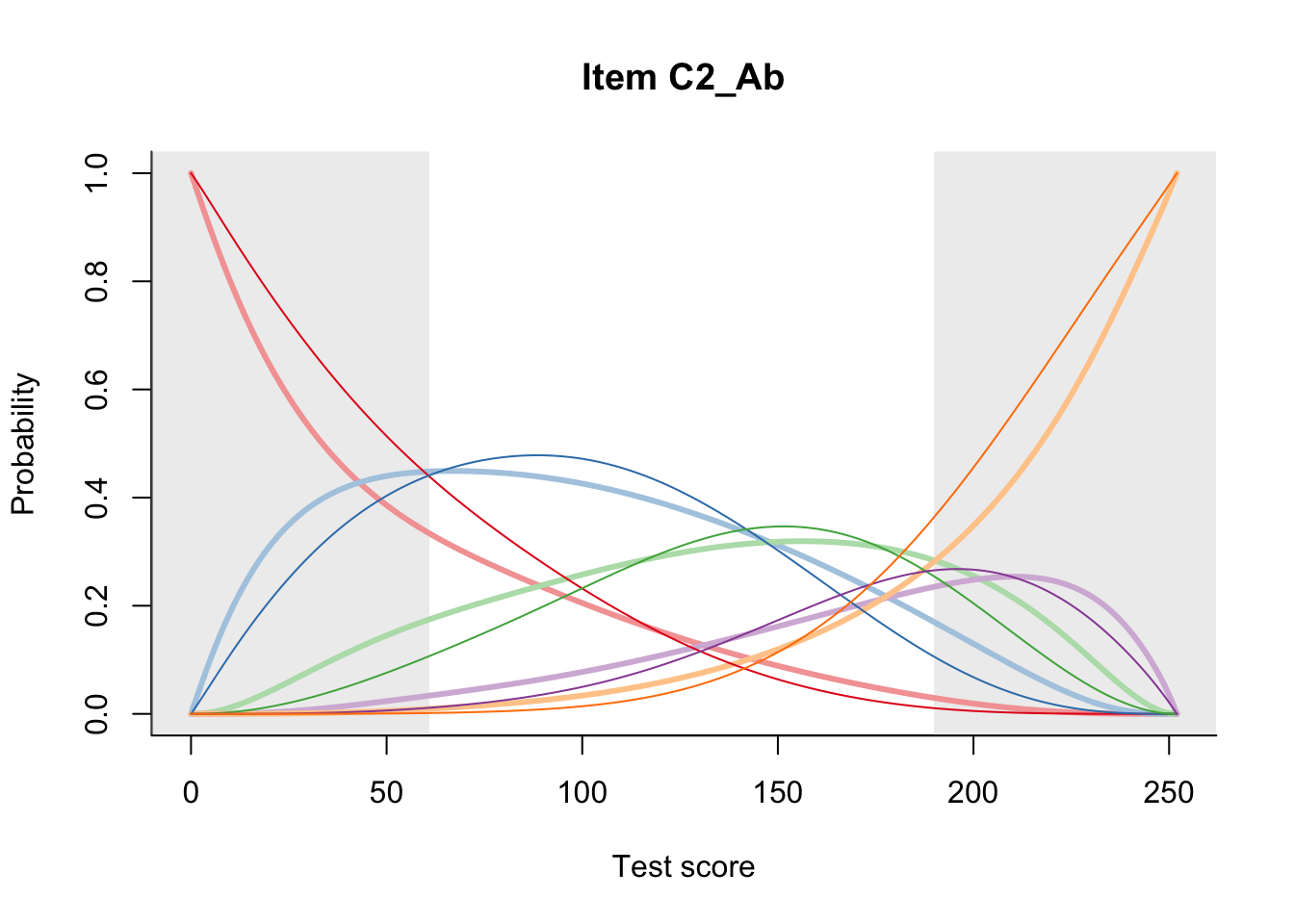
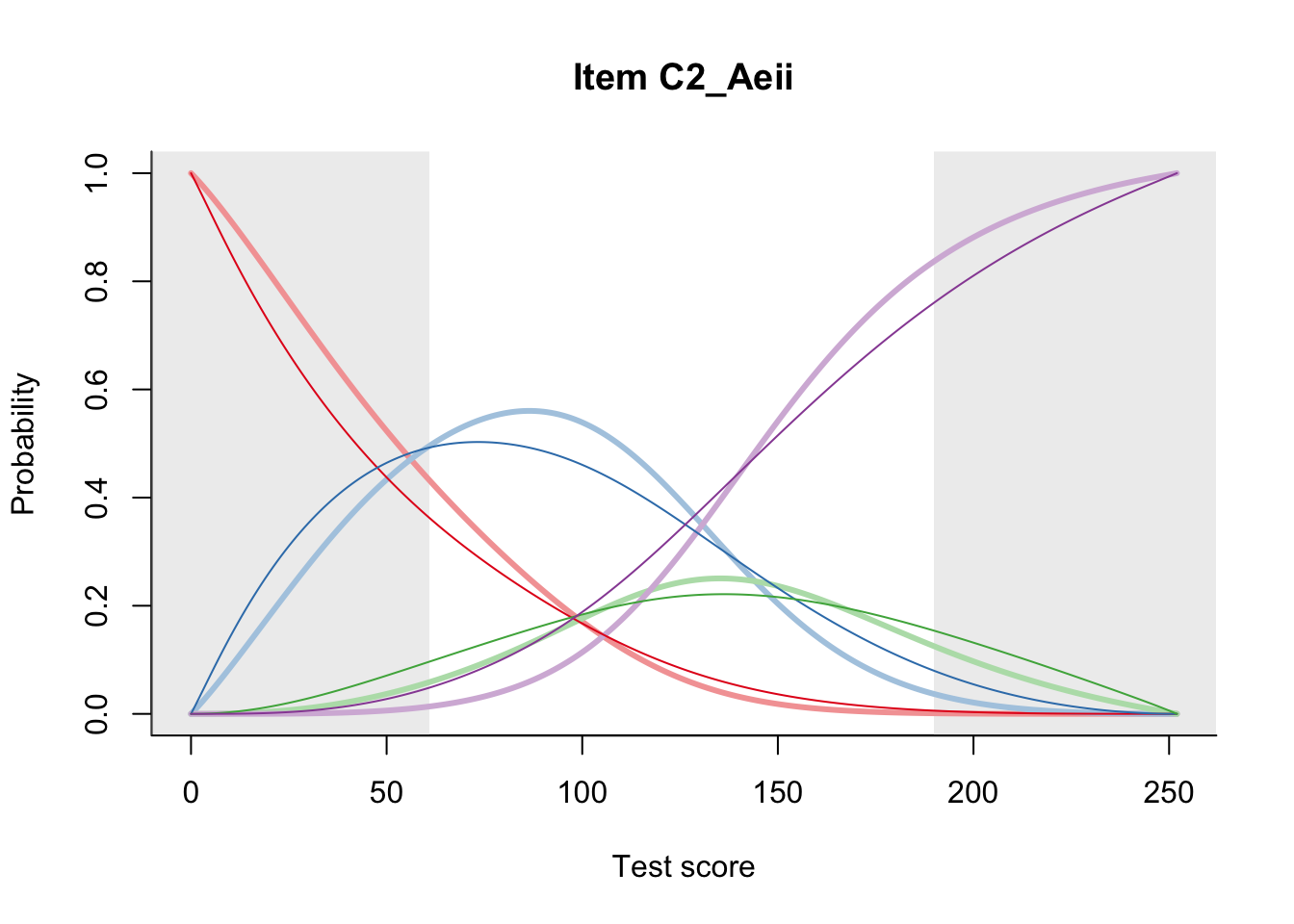
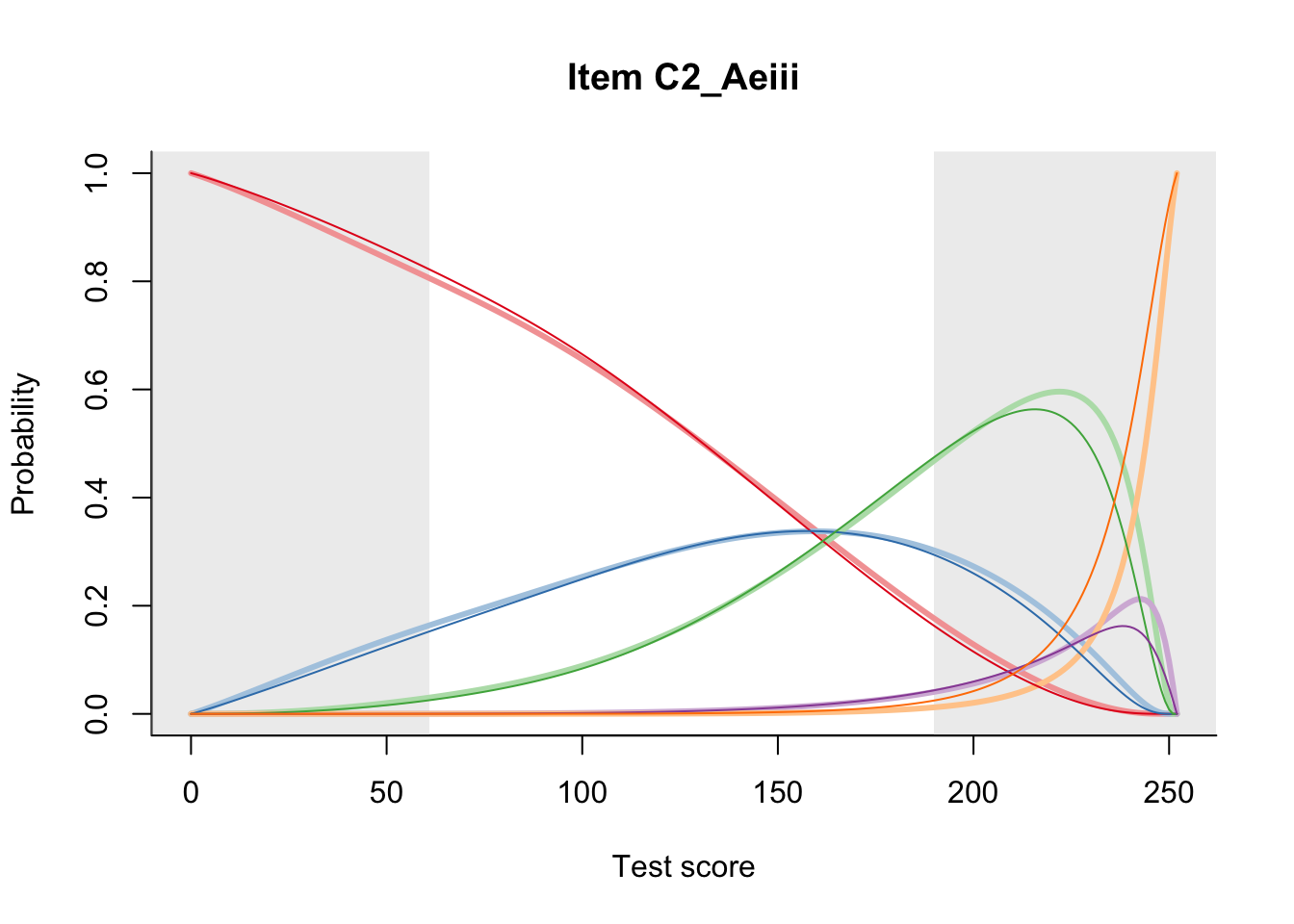
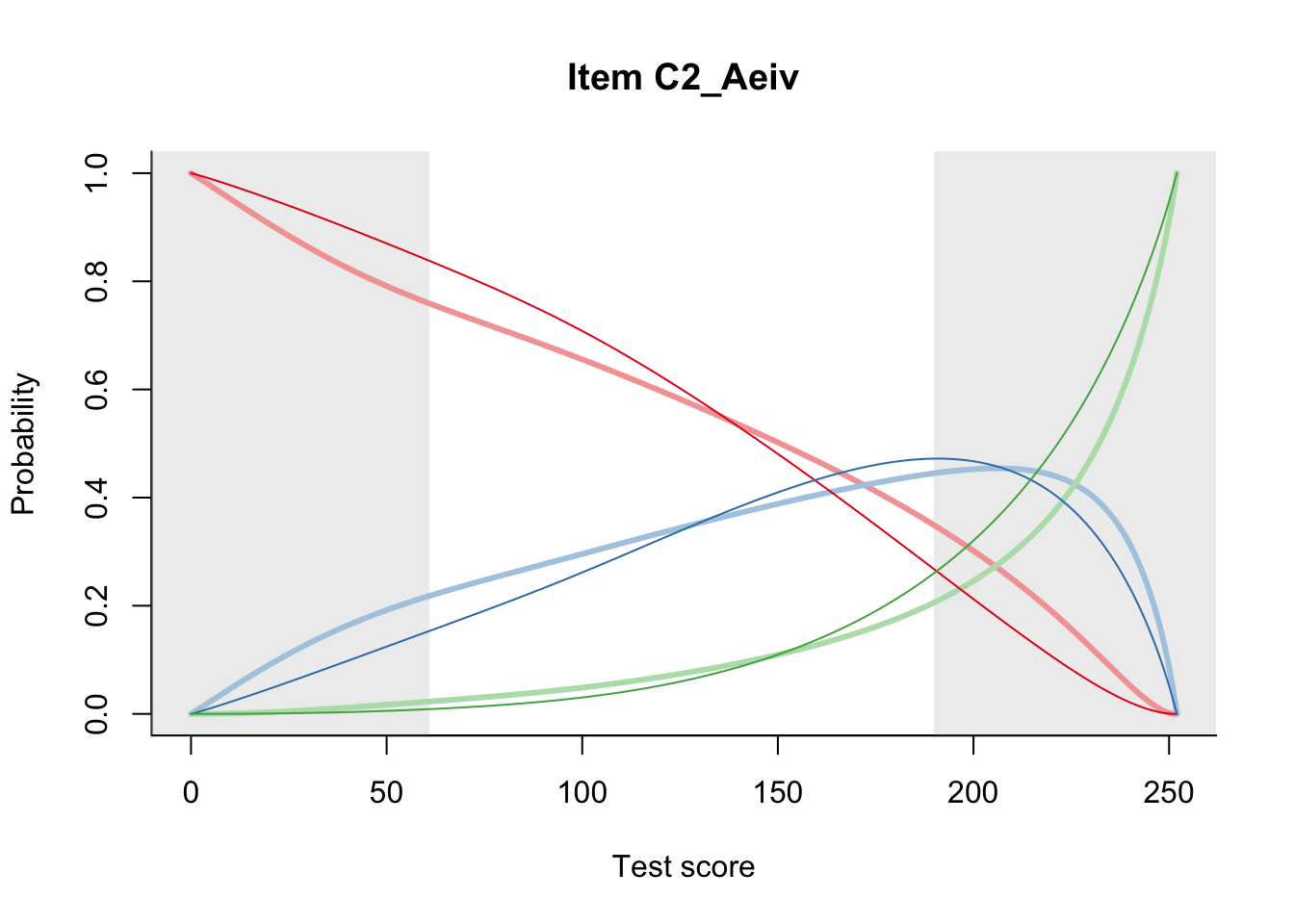
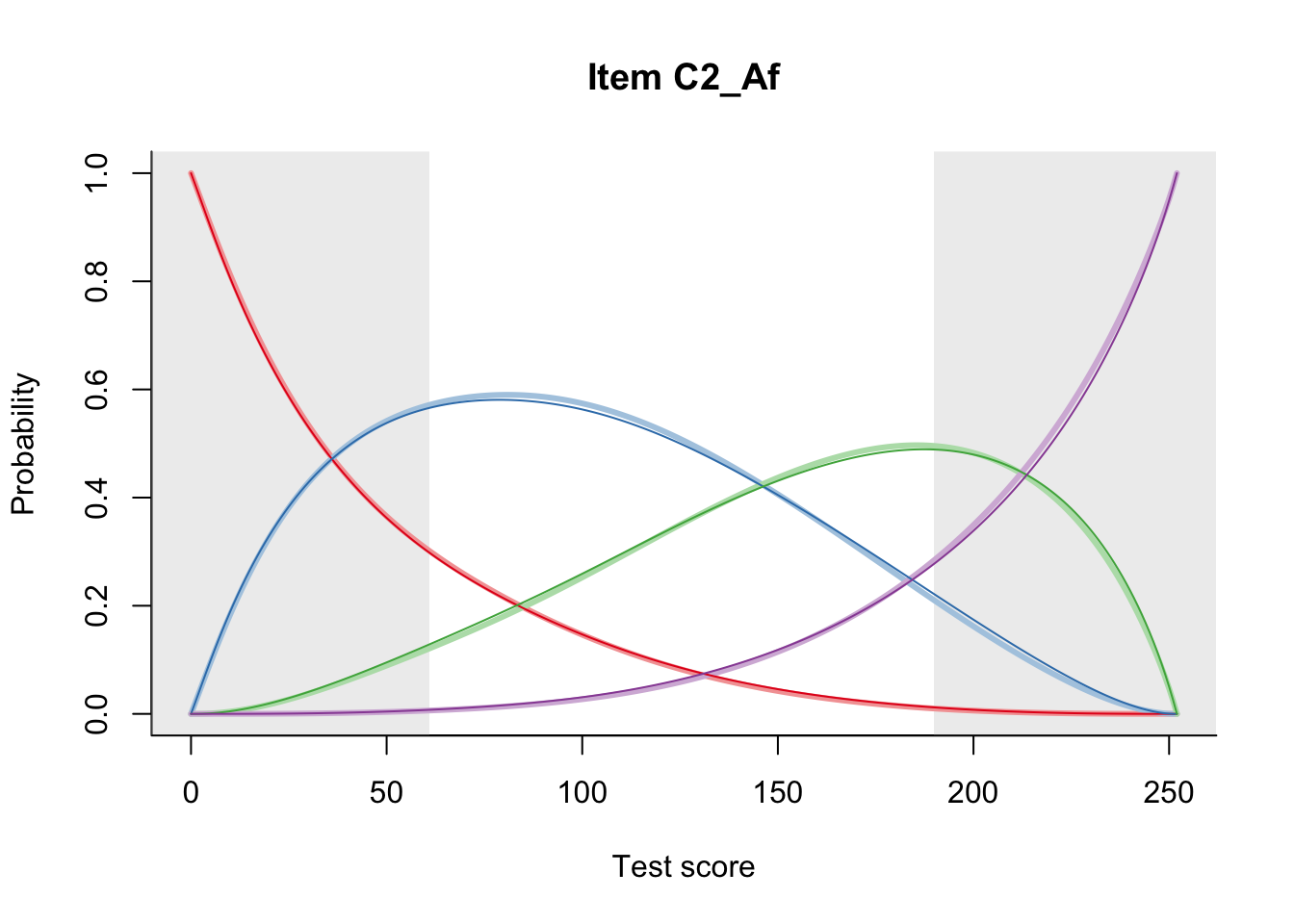
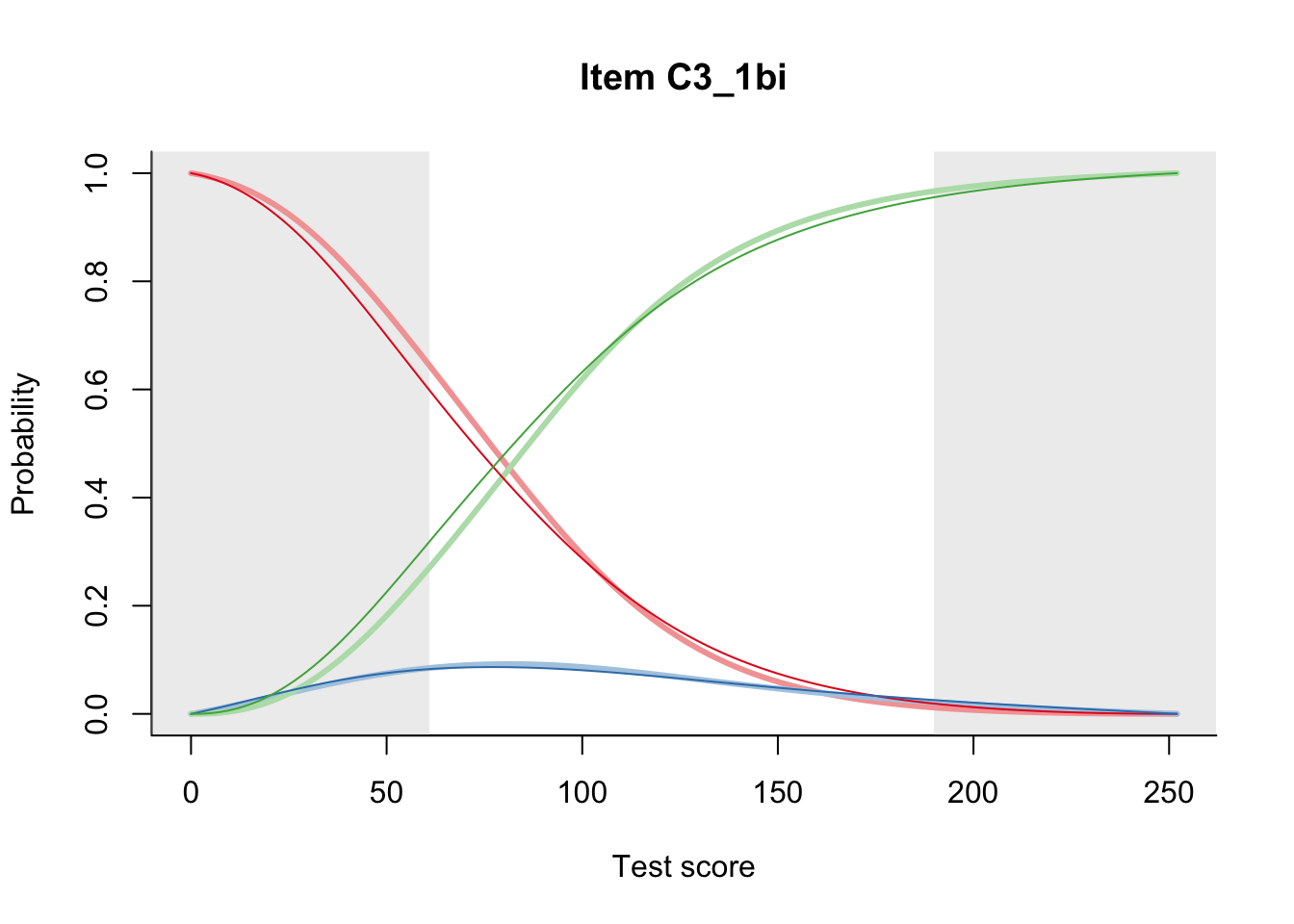
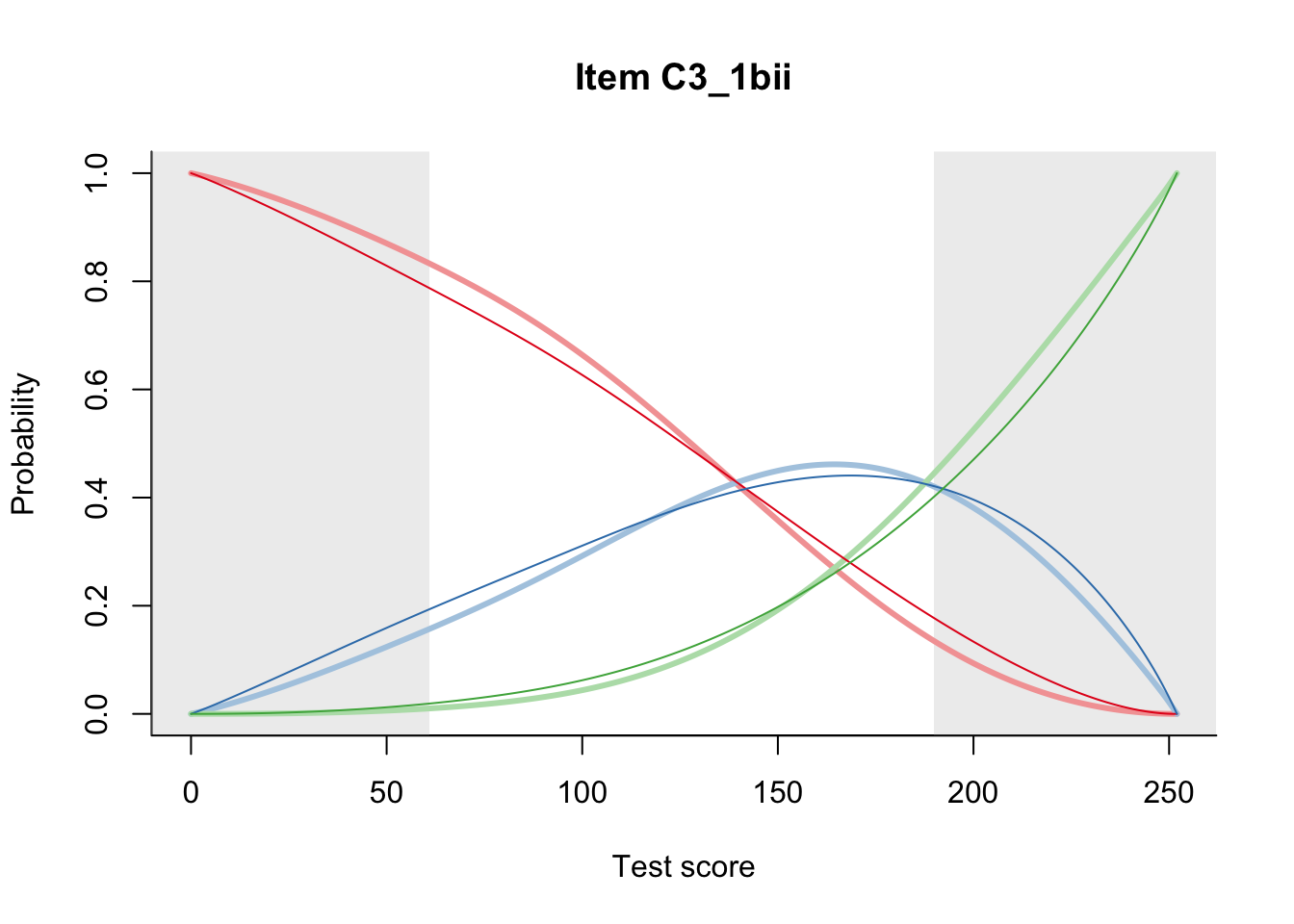
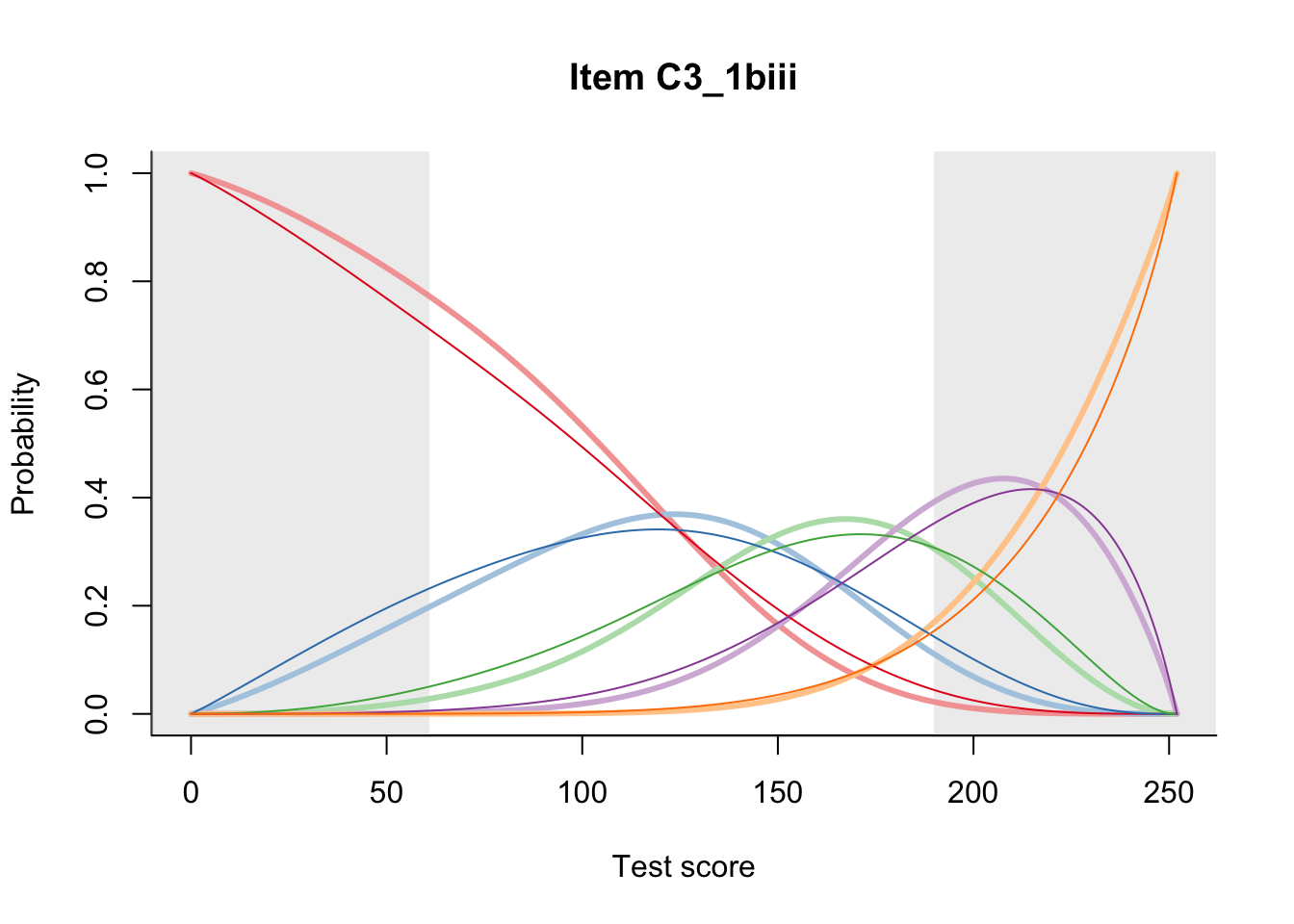
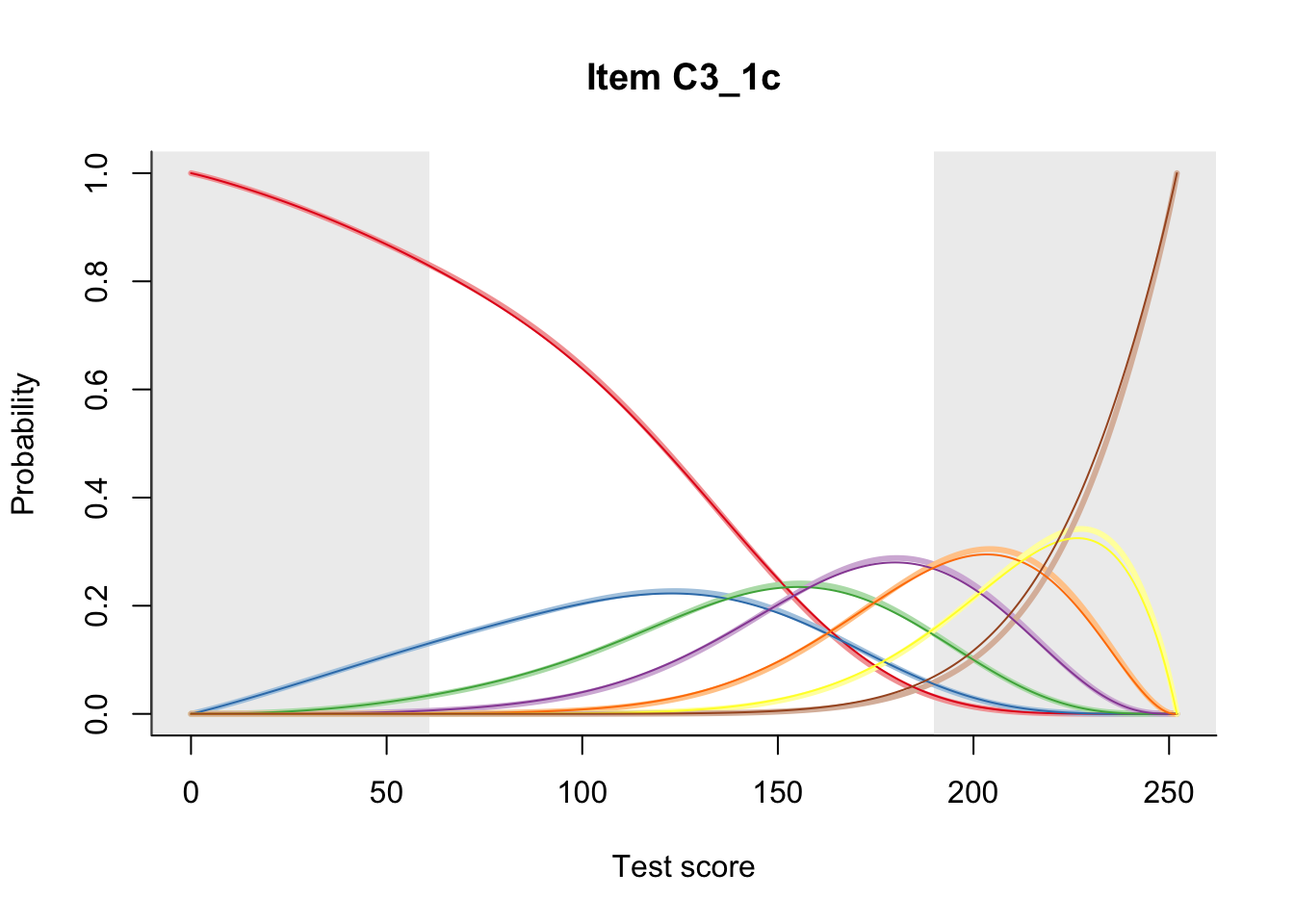
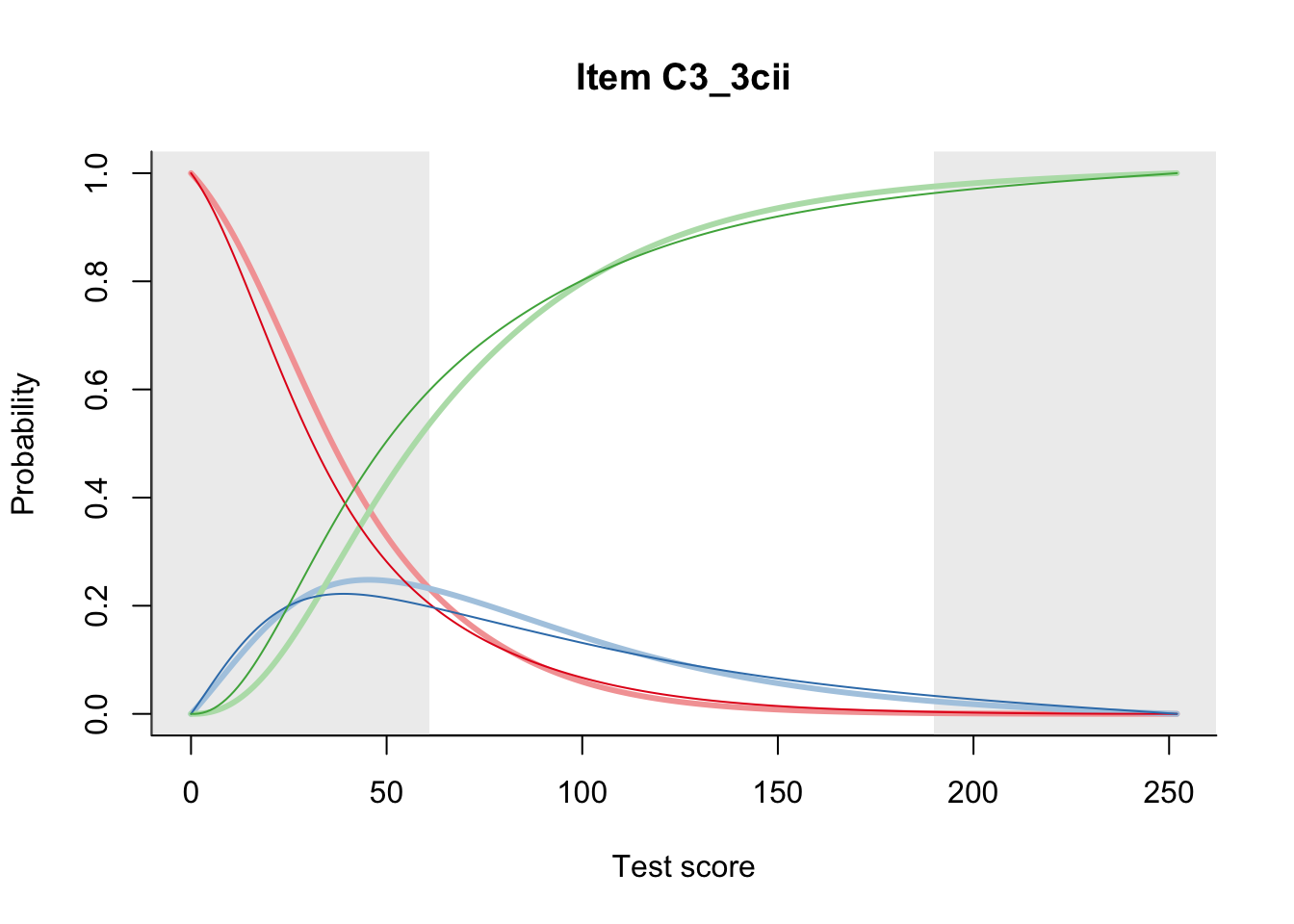
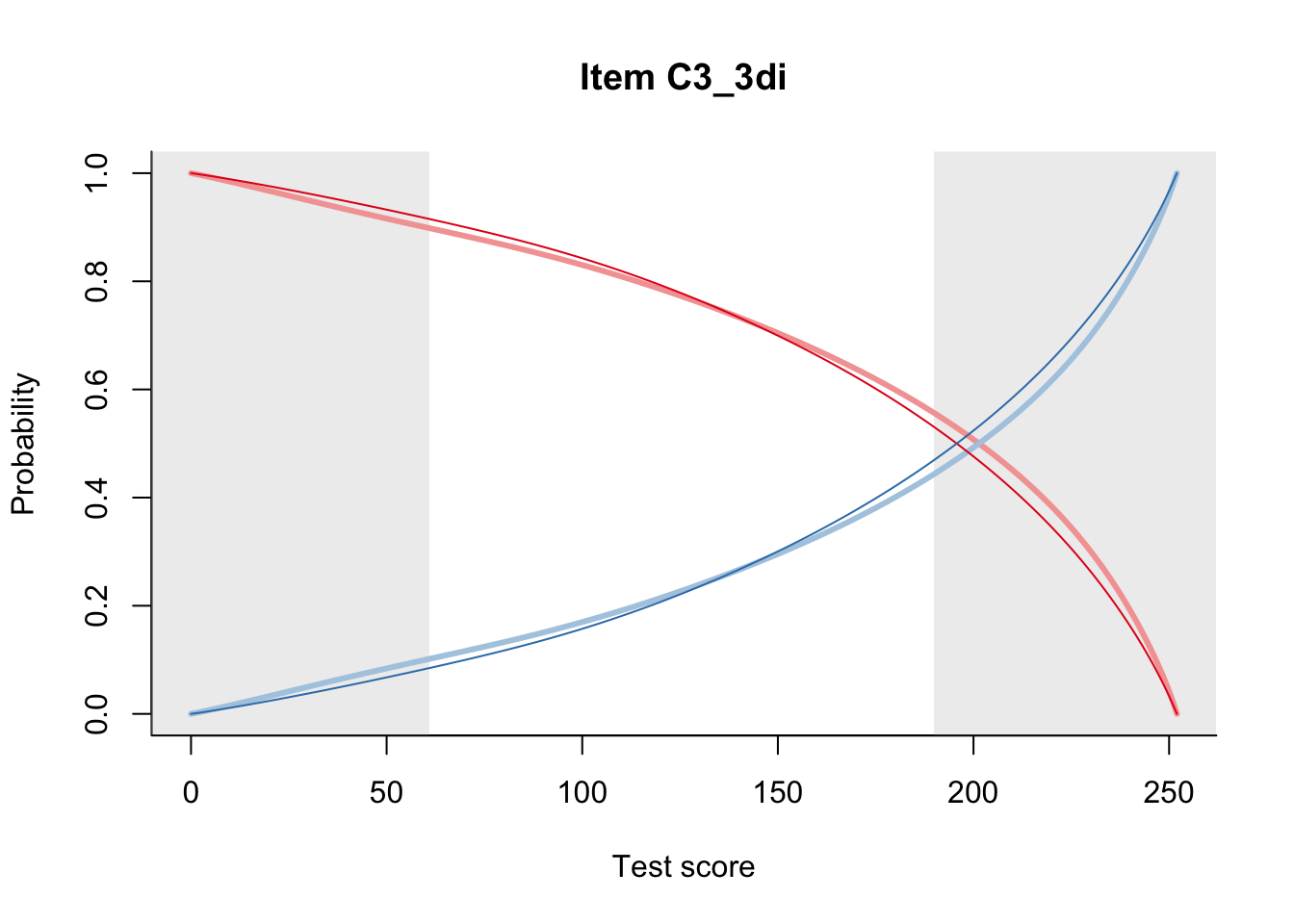
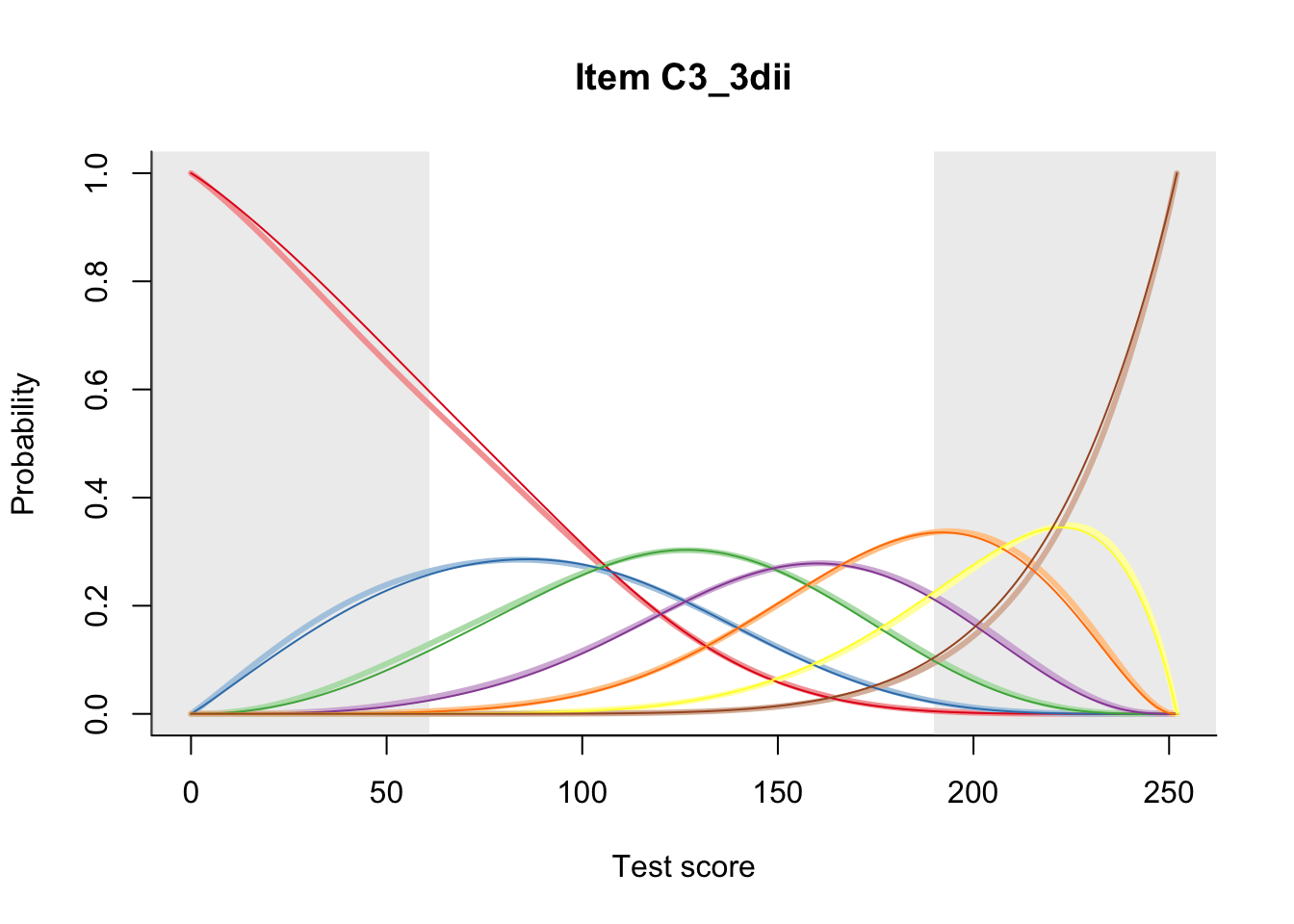
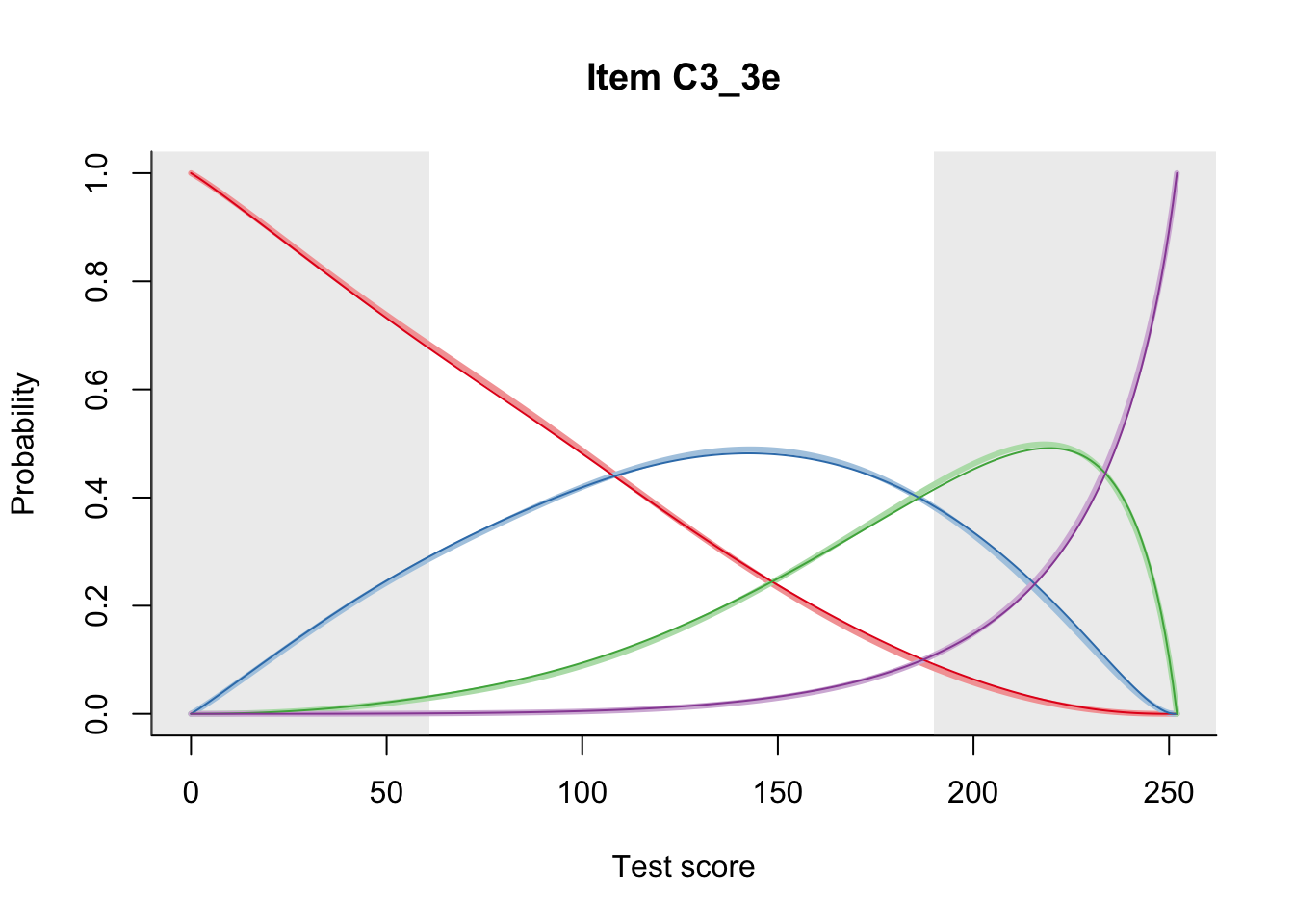
# plot(m, "C3_1ai", summate=FALSE, show.observed=FALSE)
# plot(m, "C3_1ai", summate=TRUE, show.observed=TRUE)
close_project(db)